Preparation of herbs
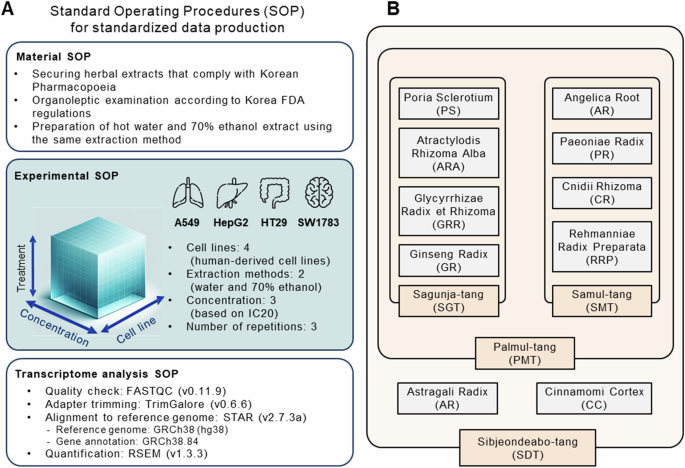
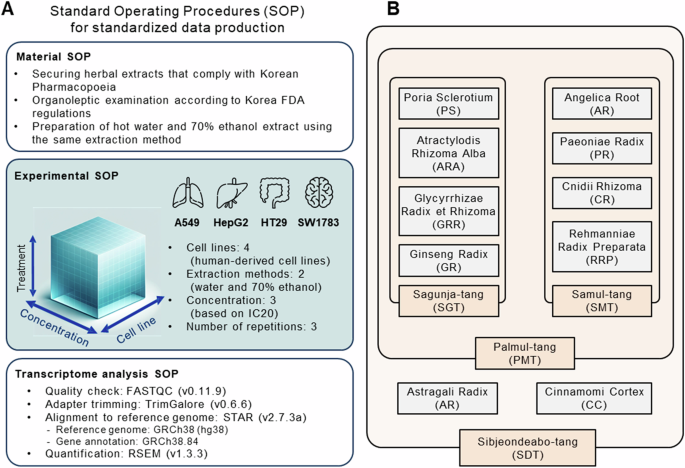
Dried medicinal crops, conforming to the Korean Pharmacopoeia requirements, have been offered by Kwangmyung-dang Medicinal Herbs Co., situated in Ulsan, Republic of Korea. These samples underwent an organoleptic examination by Dr. Choi Goya, a natural medication organoleptic examination knowledgeable appointed by the Korea Meals and Drug Administration. The identification to species degree was completed by DNA barcode area sequencing. Voucher specimens have been saved on the Korean Herbarium of Commonplace Natural Assets, inside the Natural Drugs Assets Analysis Heart, on the Korea Institute of Oriental Drugs in Naju, Republic of Korea (Desk 1). All herbs and extracts, which have been sourced from the Oriental Drugs Assets Analysis Heart (KIOM), can be found on-line at https://oasis.kiom.re.kr/herblib.
Preparation technique of scorching water and 70% ethanol extracts of herbs and THMs
Sizzling water and 70% ethanol extracts of every plant have been ready and equipped by KOC Biotech Co., situated in Daejeon, Republic of Korea. Initially, dried crops (1,000 g) have been pulverized and extracted in 10 L of scorching distilled water for 3 h utilizing a reflux extraction system (MS-DM609; MTOPS, Seoul, Republic of Korea), or in 10 L of 70% ethanol for 1 h utilizing an ultrasonication system (VCP-20, Lab companion, Dajeon, Republic of Korea) twice. The ensuing extract options have been filtered by a 5 µm cartridge filter, concentrated utilizing a rotary evaporator (Ev-1020, SciLab, Seoul, Republic of Korea), and eventually lyophilized in a freeze dryer (LP-20, Ilshin-Bio-Base, Dongducheon, Republic of Korea) to provide the ultimate extracts. These extracts have been then finely homogenized and packaged in glass bottles with desiccant silica gel. THMs have been ready by mixing and homogenizing these extracts in accordance with the composition ratios and extract yields of the person medicinal herbs, in keeping with the Korean Pharmacopoeia (Desk 2). For in vitro purposes, extracts (100 mg) have been vigorously vortexed for 30 min in 10 mL of phosphate-buffered saline (PBS; Thermo Fisher Scientific, Rockford, IL, USA) containing 2% DMSO. This combination was then sterilized by filtration by a 0.22 µm membrane to acquire a inventory resolution (10 mg/mL), which was divided into small aliquots and saved at −80 °C till their use.
Cell tradition
All cell strains have been bought from the American Kind Tradition Assortment (Manassas, VA, USA) and have been cultured in a basal medium enriched with 10% heat-inactivated fetal bovine serum, 100 IU/mL penicillin, and 100 µg/mL streptomycin, all inside a humidified incubator (Desk 3). Cell confluence ranges between 80–90% prompted the substitute of the expansion medium each 3–4 days to keep up optimum progress situations. To make sure the absence of mycoplasma contamination, the MycoAlert PLUS mycoplasma detection equipment (Lonza, Rockland, ME, USA) was employed for normal testing.
Drug therapy and complete RNA preparation for RNA sequencing (RNA-seq) evaluation
To find out the suitable therapy drug concentrations, we carried out cell cytotoxicity checks to analyze drug doses that maintained 80% cell viability (IC20s), which have been then adopted because the maximal doses for RNA-seq knowledge assortment. For medicine whose IC20s couldn’t be decided, the very best therapy concentrations have been capped at 500 µg/mL for extracts, contemplating each their solubility and relevance for medical software. To verify the affect of focus, cells have been handled with three completely different concentrations utilizing 1/5 serial dilutions, thereby exposing them to low, medium, and excessive doses. Constructive management medicine comparable to wortmannin (Sigma, W1628), LY294002 (Sigma, L9908), and Thioridazine (Sigma, T9025) have been integrated into the assay for comparative evaluation in opposition to the connectivity map (CMap) knowledge. Cells handled with a 2% DMSO/PBS resolution served because the automobile management. Someday earlier than drug administration, cells have been seeded into 6-well tradition plates with 3 mL of progress medium. Following a 24 h therapy interval, the cells have been washed with ice-cold PBS, and complete RNA was remoted utilizing QIAzol RNA isolation reagents (Thermo Fisher Scientific) in accordance with the producer’s directions.
RNA-seq knowledge technology and preprocessing
Complete RNA (over 500 ng) from every pattern was processed for the mRNA sequencing library utilizing the MGIEasy RNA directional library prep equipment (MGI Tech Co., Ltd., China), following the producer’s directions. The library focus was quantified utilizing the QuantiFluor® ssDNA System (Promega Company, WI, USA). The ready DNA nanoball was sequenced on an MGISEQ system (MGI Tech Co., Ltd., China) using 100 bp paired-end reads. The RNA-seq knowledge high quality was assessed utilizing FastQC (v0.11.9). To take away frequent MGISEQ adapter sequences, TrimGalore (v0.6.6) was utilized. Trimmed reads have been then mapped to the human reference genome meeting GRCh38 (hg38) utilizing the STAR aligner (v2.7.3a) with default settings12. Gene transcript abundance, together with anticipated learn counts and transcripts per million, was quantified utilizing RSEM (v1.3.3), with the gene annotation GRCh38.8413. The uncooked sequence knowledge (FASTQ recordsdata) and the preprocessed expression values for every gene have been deposited within the GEO beneath accession numbers GSE244687, GSE244707, GSE244694, and GSE245912.
Differential gene expression evaluation
Utilizing the gene symbols of protein-coding genes, we utilized the collapseRows perform from the WGCNA bundle (v.1.72-1)14, particularly designed to merge expression knowledge for genes represented by a number of probes. This method successfully reduces redundancy and potential noise, enhancing the readability of subsequent analyses. Moreover, the filterByExpr perform from the genefilter bundle (v.1.78.0)15 was utilized to exclude genes that failed to satisfy predetermined expression standards throughout samples. This filtering ensured that solely genes most definitely to offer dependable and related indicators have been retained for evaluation.
For analysis of every set of therapy situations—encompassing 4 cell strains, 14 herbs and THMs, two extraction strategies, and three focus ranges— we performed differential gene expression (DGE) evaluation in opposition to the corresponding management samples. This evaluation was carried out utilizing the Wald check statistic as applied within the DESeq. 2 bundle (v.1.36.0)16. Differentially expressed genes (DEGs) have been decided primarily based on a fold-change threshold of 1.5 and an adjusted P-value of lower than 0.05.
Clustering evaluation
The fold-change values derived from the DGE evaluation throughout all therapy situations have been clustered utilizing the t-distributed stochastic neighbor embedding (t-SNE) algorithm. This machine studying method, designed for dimensionality discount, excels in visualizing high-dimensional datasets, making it a worthwhile software for deciphering advanced gene expression patterns. The evaluation was performed using the Rtsne bundle (v.0.16), with the perplexity parameter set to 1017.
Comparisons with connectivity map transcriptome knowledge
Connectivity Map knowledge have been obtained from the Clue.io platform(clue.io/knowledge/CMap2020#LINCS2020). For our evaluation, we chosen degree 5 gene expression signatures with excessive reproducibility, outlined by moderated z-scores that met particular standards (distil_cc_q75 > 0.5 and pct_self_rank_q25 > 0.05), to check with our RNA-seq knowledge. The R bundle CMapR (v1.8.0) was used to govern the extent 5 GCTX file (level5_beta_trt_cp_ n720216 × 12328.gctx). Given the variance in gene expression profiling between our RNA-seq knowledge and L1000 assays9 utilized in CMap, a direct comparability between gene expression values was tough on account of distinct distributions of expression values. To navigate this, we employed gene set enrichment evaluation (GSEA) in its place technique to discover the genome-wide perturbing results of therapies comparable to wortmannin on the pathway degree18. We utilized 2,229 gene units from a number of databases—Hallmark, Biocarta, KEGG, REACTOME, PID, and Wikipathways—obtainable by MSigDB (https://www.gsea-msigdb.org/gsea/msigdb). The evaluation concerned performing GSEA on all genes, ranked in keeping with their Wald check statistics or level5 z-scores. To acquire the MSigDB gene units and conduct GSEA, we utilized the R bundle MSigDBR (v7.5.1) and FGSEA (v3.18). From the GSEA outcomes, we outlined pathway exercise rating (PAC) as ‘signal (enrichment rating) × -log10(p-value)’ worth to quantify the importance degree. PAC vectors of equal lengths (n = 2,229) have been generated for each our dataset and the CMap dataset. Subsequently, we decided the Pearson correlation coefficient to evaluate the connection between the PACs from our samples and people from CMap (Fig. 1).