1 Introduction
Metachromatic leukodystrophy (MLD) is an autosomal recessive lysosomal storage neurodegenerative dysfunction, pathologically characterised by progressive motor and cognitive dysfunction (Miyake et al., 2021; Lamichhane and Cabrero, 2023; Penati et al., 2017; Singh and Singh, 2024b). MLD is attributable to the deficiency of aryl sulfatase A (ARSA), which results in the buildup of sulfatide, the principle glycolipid concerned in stabilizing the myelin sheath equilibrium within the central and peripheral nervous programs (Sessa et al., 2016; Fumagalli et al., 2022). Sulfatide accumulation might trigger numerous pathophysiological situations, together with progressive demyelination, neuroinflammation, communication hole, astrocyte dysfunction, developmental delay, speech dysfunction, and in extreme instances it causes dying inside 5–6 years of the early onset of the illness (Miyake et al., 2021; Lamichhane and Cabrero, 2023; Fumagalli et al., 2022). Primarily based on the age of onset, MLD is categorized into three main medical sorts: late-infantile (≤30 months); juvenile, which is subdivided into early juvenile [30 months–6 years] and late juvenile [7–16 years]; and grownup MLD (≥17 years) (Miyake et al., 2021; Fumagalli et al., 2022; Chang et al., 2024). Worldwide, the prevalence of MLD is 1.4–1.8 in 100,000 or 1 in 40,000, bringing it to the loop of uncommon ailments of higher concern (Lamichhane and Cabrero, 2023; Chang et al., 2024; Shaimardanova et al., 2020).
So far, greater than 280 mutations have been reported within the ARSA gene, which poses a serious problem within the success of present therapeutic choices, together with gene remedy, hematopoietic stem cell remedy, enzyme substitute remedy, and chaperone remedy, thus making these choices pricey and, due to this fact, demanding a case-by-case strategy that leads the therapy of this uncommon illness past the attain of the bigger inhabitants (Shaimardanova et al., 2020; Kurtzberg, 2022; Eichler et al., 2022; Fernández-Pereira et al., 2021; Sevin and Deiva, 2021). One other main impediment to the success of present therapies is the blood–mind barrier (BBB) in delivering the therapeutic gene or enzyme to the goal website (Miyake et al., 2021). In distinction to present therapies, that are primarily ARSA-dependent, a brand new strategy of substrate discount remedy (SRT) may be an alternate therapeutic choice for metachromatic leukodystrophy. SRT has proven its success in numerous single-gene issues together with some key lysosomal storage issues (Mistry et al., 2023; Komada et al., 2021; Istaiti et al., 2022). SRT focuses on the event of oral medicine to deal with pathophysiological situations. Miglustat and eliglustat are the 2 FDA-approved oral medicine for Gaucher’s illness (Mistry et al., 2023; Komada et al., 2021; Istaiti et al., 2022).
In MLD, the therapeutic focus in substrate discount remedy is the event of a particular, potent, and aggressive inhibitor which targets the catalytic motion of the rate-limiting enzyme, cerebroside sulfotransferase, that’s concerned within the biosynthesis of sulfatides (Singh and Singh, 2024b; Yaghootfam et al., 2007; Li et al., 2020). So far, the event in substrate discount remedy has been on the nascent stage in MLD. Experimental knowledge with obtainable CST inhibitors are restricted, though the full-length CST sequence and cDNA can be found (Li et al., 2020). The shortage of structural info of proteins as a result of unavailability of the three-dimensional construction of CST has been a serious roadblock in initiating preliminary in silico-based drug discovery, which has nice potential in screening drug-like candidates within the shortest potential time with much less bills. The event of the three-dimensional homology mannequin of CST by our group, which used a multipronged modeling and validation strategy, is a breakthrough in SRT analysis in MLD (Singh and Singh, 2024a). This examine makes use of this mannequin for the screening of phytoconstituents from Medhya Rasayana.
Medhya Rasayana is a bunch of Ayurvedic medicinal herbs with neuroprotective results in opposition to numerous neurodegenerative ailments and identified to enhance cognitive operate and neural tissue regeneration, retard mind getting older, and stability psychological well being (Kulkarni et al., 2012; Sarokte and Rao, 2013; Rashmi et al., 2017). Medhya Rasayana contains 4 main herbs: Mandukaparni (Centella asiatica Linn.), Yastimadhu (Glycyrrhiza glabra Linn.), Guduchi (Tinospora cordifolia Miers), and Shankhpushpi (Convolvulus pluricaulis Chois) (Kulkarni et al., 2012). These herbs are wealthy in phytoconstituents with distinctive bioactive properties. This examine focuses on figuring out potent and particular bioactive compounds as inhibitors in opposition to CST by means of multipronged in silico research. Excessive-throughput digital screening is the commonest and dependable technique used to determine the potent and particular inhibitors and potential lead molecules from the varied dataset of small molecules at low price and fast tempo utilizing the current development within the discipline of bioinformatics (Singh et al., 2018; Singh et al., 2017; Yasuo and Sekijima, 2019). With this angle, the current examine opens the door for a lot of futuristic research to attain a marketed oral drug for metachromatic leukodystrophy that might convey this hereditary illness inside the attain of a potential cost-effective therapy loop.
2 Methodology
2.1 Sources
The computational examine was executed utilizing the high-performance super-computing facility, PARAM Shivay, put in on the Indian Institute of Expertise, Banaras Hindu College, Varanasi, India, with a capability of 837 TFLOPS with Intel(R) Xeon(R) Gold 6148 CPU @2.40 GHz and 40 CPUs per node. This examine used key software program packages together with AutoDock 4.2, GROMACS 2023, Origin 2024, Chimera 1.17.3, PyMOL, Discovery Studio visualizer, VMD, and Bio3D instrument with R package deal and key webtools together with PharmMapper, pkCSM, SwissADME, and ProTox 3.0.
2.2 Protein structural evaluation and receptor grid era
For screening of bioactive phytoconstituents, the 3D mannequin of CST developed in our earlier examine was used as a receptor. The mannequin contains 69–336 amino acid residues of the full-length protein with 423 residues and covers the whole catalytic area of the protein with a sulfuryl acceptor substrate (galactocerebroside)-binding website and a sulfuryl donor co-substrate (PAPS)-binding website on a linear horizontal aircraft (Singh and Singh, 2024a; Singh and Singh, 2024b). The mannequin protein was processed utilizing AutoDock 4.2 by the removing of water molecules and heteroatoms and the addition of hydrogen atoms with correct task of atom kind and Gasteiger cost to generate a PDBQT file of the protein. A grid field of 90 × 90 × 90 Å and spacing of 0.253 Å across the protein lively website was generated contemplating the important thing residues LYS82, HIS84, LYS85, HIS141, PHE170, TYR176, PHE177, TYR203, and ARG202, and a grid file (.gpf) was created.
2.3 Ligand preparation
We used a set of compounds from the Indian Medicinal Crops, Phytochemistry, And Therapeutics (IMPPAT 2.0) on-line database (Vivek-Ananth et al., 2023; Mohanraj et al., 2018). The 3D (.mol2) buildings of 81 compounds from C. asiatica Linn (Mandukaparni), 310 from G. glabra Linn (Yastimadhu), 52 from T. cordifolia Miers (Guduchi), and 18 from C. pleuricaulis Chois (Shankhpushpi) had been transformed to .pdbqt recordsdata after power minimization and assigning correct atom sorts utilizing AutoDock Raccoon, a digital screening file preparation instrument (Forli et al., 2016).
Following the preparation of the protein, ligands, and grid recordsdata, AutoDock Raccoon was additional used for the preparation of docking (.dpf) recordsdata for every ligand and the following association of all recordsdata in a single separate folder for every protein–ligand advanced with the era of a single digital screening script file (.sh) for performing molecular docking-based digital screening of all ligands concurrently below the Linux setting. The parameters used for the era of .dlg recordsdata for docking run had been 100 GA run, a inhabitants dimension of 300, most variety of generations of 27,000, and most variety of evaluations of 25,000,000. AutoDock utilized the Lamarckian genetic algorithm and a gradient-based native search methodology for protein–ligand interactions. The pKi and ligand effectivity are two main parameters for assessing the binding potential of ligands within the lively website. The most effective-docked conformation was chosen, processed utilizing customized Python scripts, and visualized utilizing .pdb visualization instruments together with PyMOL and Discovery Studio Visualizer to investigate protein–ligand interactions and ligand-binding patterns within the lively website.
2.4 In silico drug-likeness properties, ADME, and toxicity evaluation
The chosen high hits based mostly on the binding rating and variety of conformations within the largest cluster had been subjected to pharmacokinetic property evaluation utilizing pkCSM, SwissADME, and ProTox webservers (Banerjee et al., 2018; Daina et al., 2017; Pires et al., 2015). The canonical Simplified Molecular Enter Line Entry System (SMILE) of the chosen compounds was used because the entry knowledge in these servers for his or her pharmacokinetic and drug-likeness property evaluation by means of adsorption, distribution, metabolism, and excretion together with toxicity research.
2.5 Molecular dynamics simulations
All atom molecular dynamic (MD) simulations had been carried out utilizing the GROMACS 2023.1 software program package deal utilizing the CHARMM27 all-atom additive power discipline. For the simulation, a dodecahedron simulation field was created with a minimal distance of 1.2 Å from the field edge, and periodic boundary situations had been utilized to reduce the sting impact. Every field was solvated with the TIP3P water solvation mannequin, and the costs on the system had been then neutralized by the addition of chloride (Cl−) ions, and thereafter, the solvated system was energy-minimized utilizing the steepest descent algorithm. The energy-minimized system was then equilibrated to 1,000-ps (1 ns) NVT simulation with a time step of two fs at 300 Okay. Subsequent, the system was equilibrated to 1,000-ps NPT simulations with a time step of two.0 fs at 300 Okay. The LINCS algorithm was used to constrain the bond lengths. The ultimate MD simulation run was carried out for 100 ns, and trajectories had been analyzed utilizing totally different MD parameters together with root imply sq. deviation (RMSD), root imply sq. fluctuation (RMSF), radius of gyration (Rg), solvent-accessible floor space (SASA), hydrogen bonds, principal part evaluation (PCA), and dynamic cross-correlation matrix (DCCM) evaluation of the protein–ligand complexes.
2.6 Cross-target prediction
The PharmMapper server was used to determine a variety of targets utilizing an revolutionary reverse pharmacophore mapping strategy (Wang et al., 2017; Liu et al., 2010). The most effective mapping poses of the submitted molecule (.mol2) had been aligned in opposition to all goal proteins obtainable in PharmTargetDB. The algorithm used to carry out the reverse pharmacophore matching protocol contains a sequential mixture of triangle hashing (TriHash) and genetic algorithm (GA) optimization (Liu et al., 2010). Primarily based on the calculated highest fit-score (cutoff >5.0) between the small compound and the pharmacophore fashions, the possible protein targets had been ranked. It was crucial to verify the disease-causing potential of the targets.
3 Outcomes
3.1 Molecular docking-based digital screening
Digital screening is a computational strategy used to display screen libraries of compounds obtainable in databases in opposition to the goal protein to determine a possible drug candidate for a focused illness (Singh et al., 2018; Singh et al., 2021; Zare et al., 2024; Pirolli et al., 2023). Within the CST-led analysis to develop substrate discount remedy for metachromatic leukodystrophy, the state-of-the-art in silico strategy could possibly be essentially the most promising and directional methodology for future research. Towards this strategy, the event of a homology mannequin of the CST protein utilizing numerous computational algorithms is a vital step (Singh and Singh, 2024a). Within the current examine, this 3D mannequin was utilized for screening particular and potent phytoconstituents of 4 main herbs of Medhya Rasayana. Out of a complete of 461 bioactive compounds, 81 compounds from C. asiatica Linn., 310 from G. glabra Linn., 52 from T. cordifolia Miers, and 18 from C. pleuricaulis Chois had been screened in opposition to CST, that are detailed in Supplementary Tables S1–S4. Within the preliminary degree of screening, utilizing the bottom free power of a binding cutoff of ≤ −7.5 kcal/mol and the variety of conformation cutoff of ≥75 within the largest conformation cluster, the highest 15 compounds had been chosen, i.e., 3 from C. asiatica and 12 from G. glabra. Compounds belonging to T. cordifolia and P. Chois had been discovered to be much less potent and fewer particular towards CST and, therefore, weren’t thought-about for additional examine. The docking rating of those high 15 compounds falls between −7.57 and −10.32 kcal/mol. With a ligand effectivity of ≤0.19 per non-hydrogen atom, the highest 15 compounds had been thought-about promising for drug-likeness property evaluation. Other than α- and β-carotene, all 13 compounds strictly adopted the Lipinski rule of 5 parameters when it comes to molecular mass (<500), variety of hydrogen bond donors (<5), hydrogen bond acceptors (<10), variety of rotatable bonds, and topological floor space (TPSA). Regardless of failing on the Lipinski rule of 5 parameters, α- and β-carotene had been thought-about for additional research due to their wider therapeutic potential in neurodegenerative ailments, and these molecules have already been examined within the human physique (Banerjee et al., 2023; Abrego-Guandique et al., 2023; Hira et al., 2019; Narisawa et al., 1996). Moreover, these compounds confirmed good lipophilicity with a log p-value > 0, which is crucial for compounds to cross the lipophilic membrane and, thus, strengthen the motion of those molecules to the goal website. Subsequently, all 15 hits had been thought-about for absorption, distribution, metabolism, and excretion (ADME) and toxicity evaluation to get rid of false positives from this record, which is supplied in Table 1.
3.2 Absorption, distribution, metabolism, excretion, and toxicity profiling
After the administration of the drug within the physique, it goes by means of absorption, distribution, metabolism, and excretion processes. On this course of, the drug interacts with fascinating or undesirable targets and causes a pharmacological influence (Zhang and Tang, 2018). Thus, the bioavailability of a drug is dependent upon the speed of absorption, metabolism, and transportation of the drug to the goal website (Zhang and Tang, 2018; Stielow et al., 2023). Human intestinal absorption and lipophilicity of compounds are different essential parameters to be thought-about to make sure the simple diffusion of compounds. The cytochrome p450-based drug response to physique metabolism displays genetic variability and performs a crucial position within the cleansing of medication and homeostasis (Zhao et al., 2021). On this examine, CYP2C9, CYP2D6, and CYP3A4 inhibitors had been used to investigate the drug response on the metabolism stage. Renal OCT2 is a renal transporter that performs an essential position in figuring out the renal clearance or deposition of medication (Zou et al., 2021; Wright, 2019). Besides few, many of the chosen compounds confirmed good intestinal absorption capability, together with physique metabolism and renal excretion potential. Since MLD is a mind dysfunction, the main deciding parameters for the potential drug candidates had been BBB permeability, which screened 4 compounds—IMPHY012226 (18alpha-glycyrrhetinic acid), IMPHY012473 (lupeol), IMPHY011609 (alpha-carotene), and IMPHY011707 (beta-carotene)—with appreciable blood–mind barrier permeability. With a view to choose a possible lead molecule, in silico toxicity examine turns into one other essential criterion because of its accuracy, accessibility, and rapidity in preclinical-level screening and offering a possible lead scaffold for additional optimization (Raies and Bajic, 2016; Hemmerich and Ecker, 2020; Yang et al., 2018). pkCSM and ProTox 3.0 are freely obtainable in silico toxicity examine servers that use the SMILE of every compound to judge numerous toxicity parameters together with Ames mutagenicity, hepatotoxicity, and cytotoxicity (Banerjee et al., 2018; Pires et al., 2015). These 4 phytoconstituents belonged to class “4” of the ProTox-predicted toxicity class based mostly on their LD50 vary between 560 and a pair of,000 mg/kg, suggesting that the chosen 4 compounds are primarily unhazardous. Table 2 supplies the small print of ADME and toxicity evaluation of the highest 15 hits, and Figure 1 reveals the structural particulars of the perfect 4 compounds screened by means of ADME and toxicity evaluation.
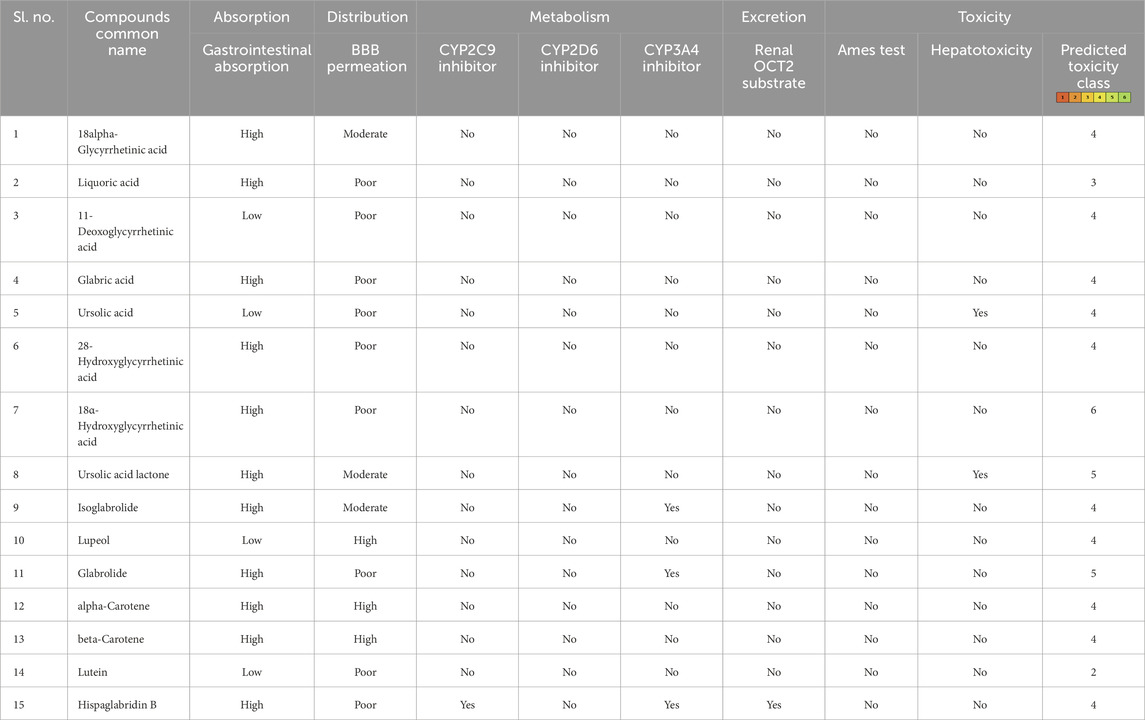
Desk 2. Pharmacological profile of the highest 15 ligand molecules that had been derived from pkCSM, SwissADME, and ProTox webservers.
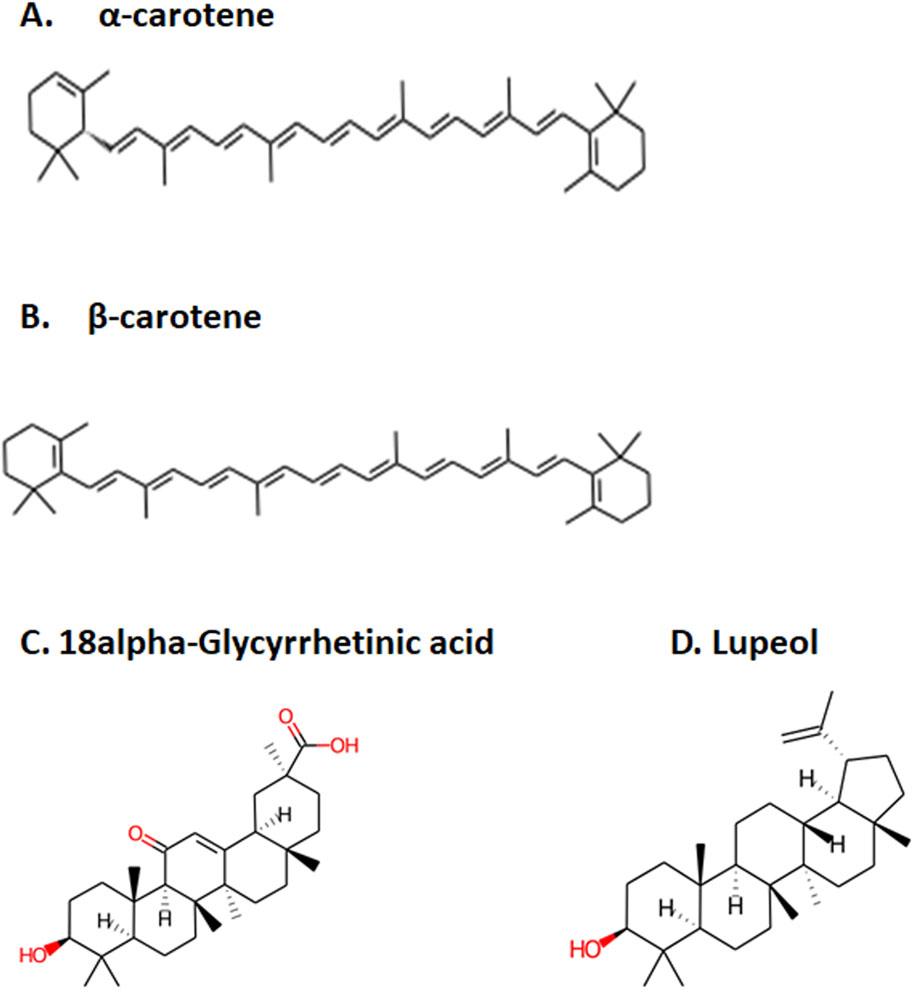
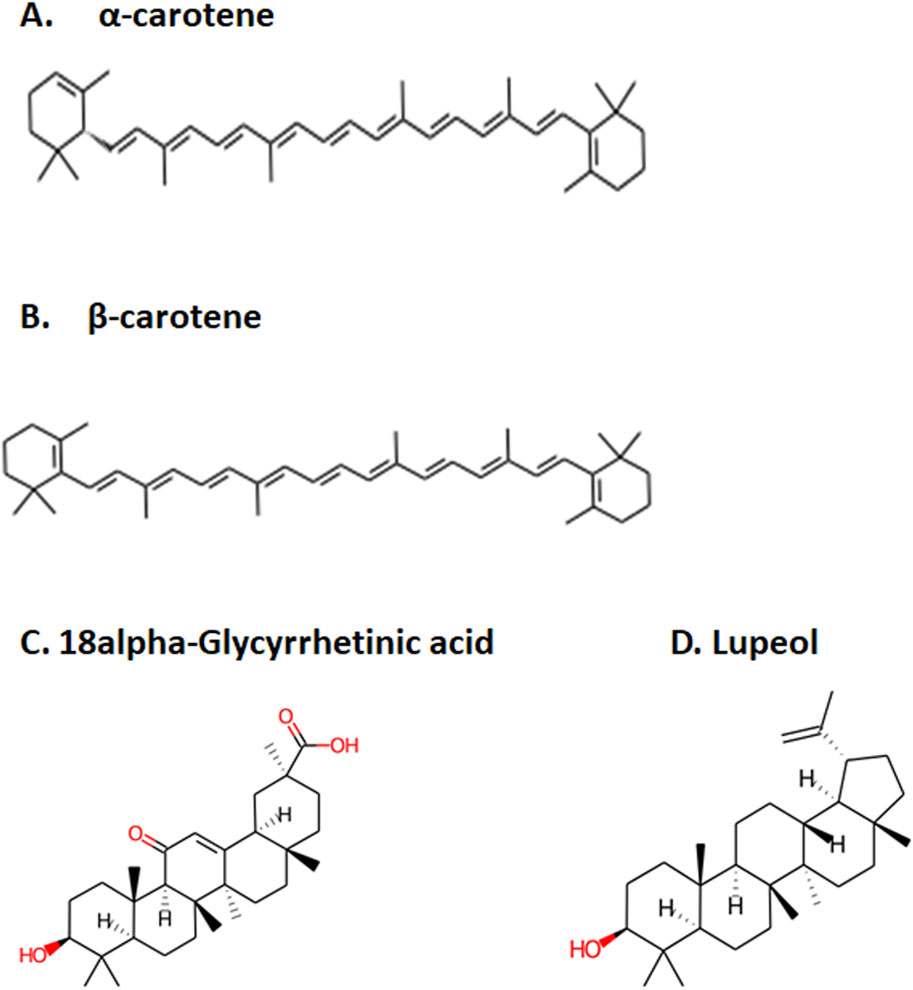
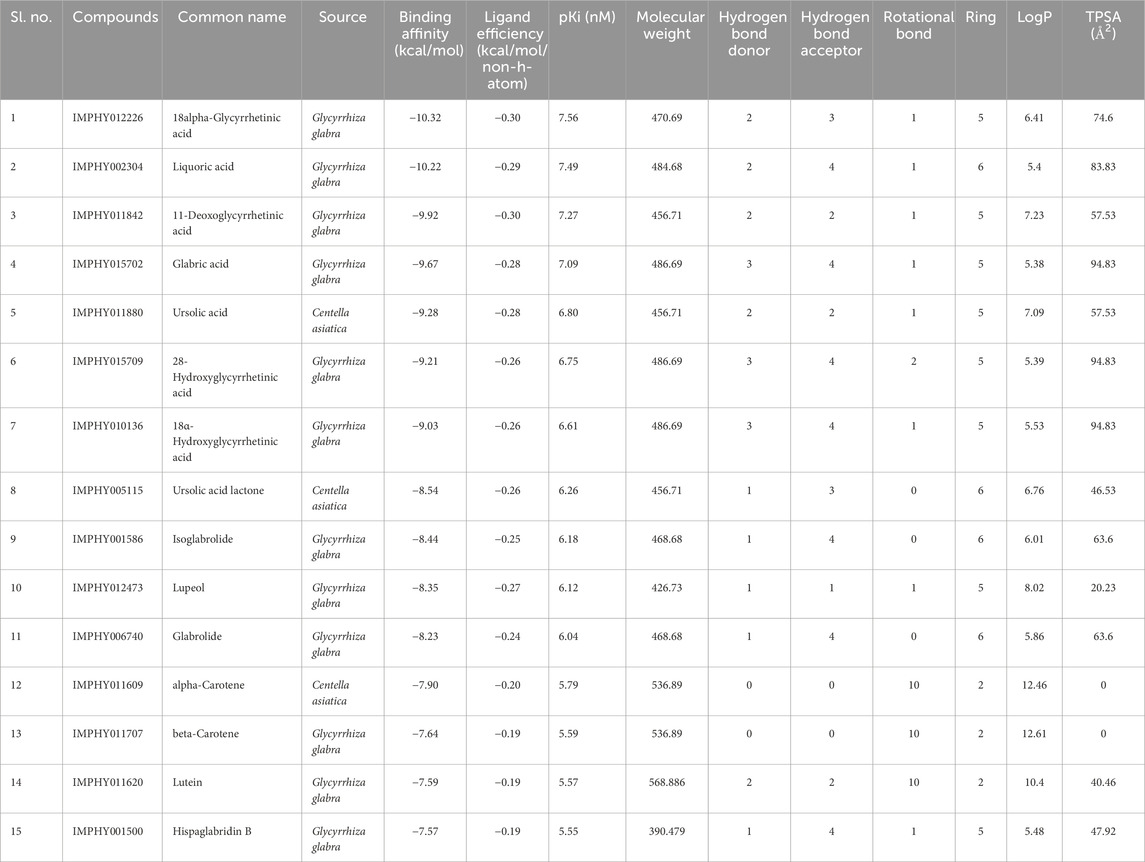
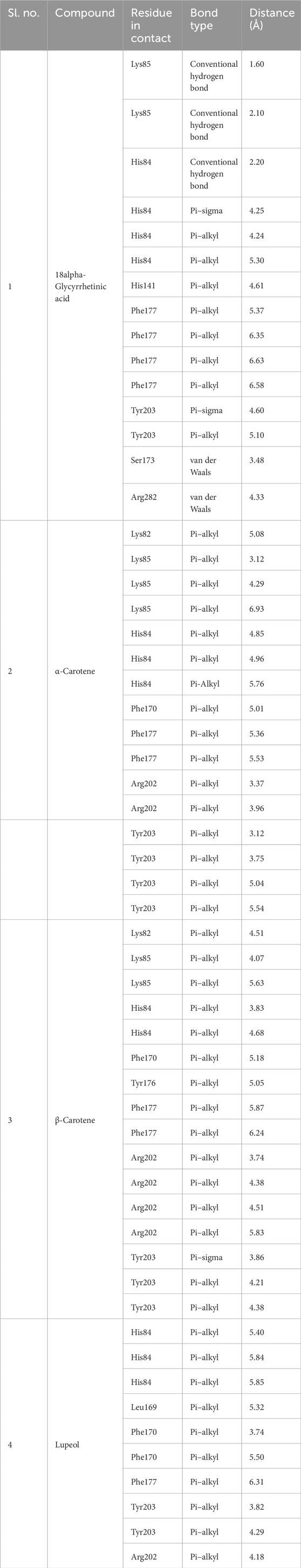
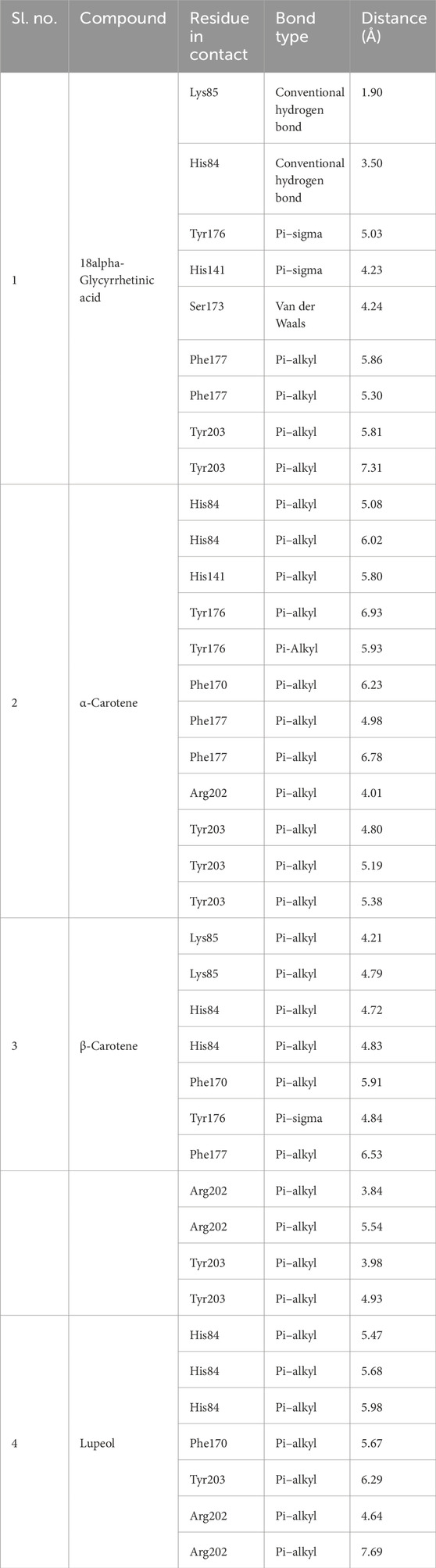
Determine 1. Prime 4 chosen compounds. (A) α-carotene. (B) β-carotene. (C) 18alpha-glycyrrhetinic acid. (D) lupeol.
3.3 Pre-MD/docking-based protein-ligand interplay pose evaluation
Protein–ligand interplay evaluation was crucial to acquire an perception into the binding sample of compounds within the substrate-binding website of the CST protein. The substrate-binding website contains a left-end polar website dominated by Lys85, Phe170, Lys82, and Ser88; a center half flanked on each side by His84 and His141; and an fragrant website on the proper finish dominated by Tyr203 and Phe170. With the bottom free binding power of −10.32 kcal/mol and 98 comparable conformations within the largest cluster, 18alpha-glycyrrhetinic acid was discovered to be essentially the most potential ligand with the very best binding affinity among the many 4 within the binding pocket of the CST protein. The compound consists of 5 steroidal rings with a carboxylic group hooked up at one finish and a hydroxyl group on the different finish. These ends decided the orientation of the compound towards the polar website based mostly on the diploma of polarity of the carboxylic group, which interacted with Lys85 by hydrogen bonding. One oxygen atom was hooked up by a double bond on the third ring in the midst of the compound positioned on the center of the lively website and was flanked on each side by His84 and His141 and shaped a hydrogen bond with His84. On the proper facet of the binding pocket, methyl teams positioned on the rightmost ring of the compound interacted with the residues Tyr203 and Phe177 through pi–sigma and pi–alkyl bonds, respectively (Figure 2I). Each α-carotene and β-carotene complexes shared similarity within the interplay sample within the CST-binding pocket. α-Carotene and β-carotene are isomers, differing within the place of the double bond on the ring at one finish oriented towards the polar website within the binding pocket. Lys82 and Lys85 performed a crucial position within the interplay with the isomeric fragment of those compounds. In α-carotene, the methyl group on the C2 place was oriented towards Lys85 and, thus, interacted with it, whereas in β-carotene, the methyl group on the C2 place was oppositely oriented and away from Lys85. Within the CST–β-carotene advanced, the 2 methyl teams on the C6 place had been oriented towards Lys85 and interacted through the pi–alkyl interplay, whereas within the CST–α-carotene advanced, the methyl teams on the C6 carbon had been positioned away from Lys85 and interacted with Lys82. The aliphatic chain between the 2 fragrant rings interacted in an almost comparable trend in each complexes with key residues Phe177, Tyr203, phe170, and His84, whereas Arg202 interacted with the fragrant ring on the appropriate facet of the binding pocket (Figures 2II, III). In distinction to the opposite three compounds that broadly occupied the lively website core, within the CST–lupeol advanced, the ligand predominantly occupied the appropriate facet of the binding pocket, which is likely to be because of its small dimension and the lacking of a polar carboxylic group at one finish because it was in 18alpha-glycyrrhetinic acid. The shortage of occupancy of the polar website by lupeol is because of its five-carbon nonpolar ring that interacted strongly on the fragrant website of the binding pocket (Figure 2IV), whereas the six-membered polar ring in 18alpha-glycyrrhetinic acid stretched the compound towards the polar website and, thus, was strongly accommodated within the lively website (Figure 2I). The main points of the interplay sorts are given in Table 3. To additional perceive the protein–ligand interplay below a dynamic setting, all 4 compounds had been thought-about for molecular dynamic simulations.
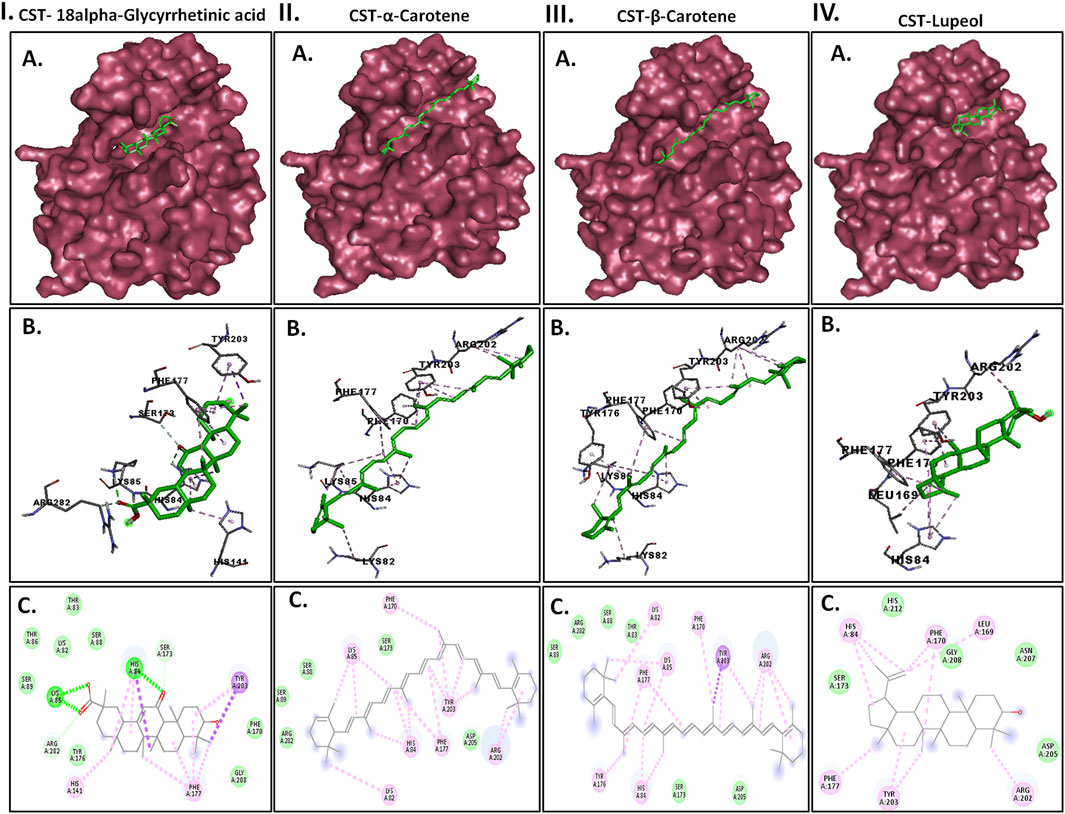
Determine 2. Interplay of the highest 4 docked complexes, together with (A) floor view (higher panel), (B) 3D view (center panel), and (C) 2D view (decrease panel) of the protein–ligand interplay on the substrate-binding website of CST protein in (I) CST–18alpha-glycyrrhetinic acid, (II) CST–α-carotene, (III) CST–β-carotene, and (IV) CST–lupeol complexes. Within the 2D view, typical hydrogen bond, pi–sigma, pi–alkyl, and van der Waals interactions are depicted in inexperienced, violet, pink, and lightweight inexperienced, respectively.
3.4 Molecular dynamic simulation
MD simulation is a computational strategy used to investigate and optimize the general stability of the protein–ligand complexes below atomistic simulation situations with a dynamic aqueous setting (Guterres and Im, 2020; Kurniawan and Ishida, 2022; Khan et al., 2016). MD simulation supplies a cumulative concept concerning the motion of each atom or atom within the protein over the simulation time span to review essential organic processes, together with the influence of ligand binding on the general protein dynamics and the way in which the macromolecule responds on the atomic degree with the binding or unbinding of the ligand (Alrouji et al., 2023; Hollingsworth and Dror, 2018). On this examine, to grasp the in-depth dynamic conduct of the protein–ligand interplay, MD simulation was carried out for a time span of 100 ns to judge the power and stability of the protein–ligand complexes by means of trajectory evaluation with numerous parameters together with RMSD, RMSF, Rg, SASA, and hydrogen bonding. We additionally carried out principal part evaluation, free power panorama evaluation, and dynamic cross-correlation evaluation to grasp the dominant motions answerable for the binding sample and stability of the protein–ligand advanced.
3.4.1 Structural deviation and adaptability with RMSD and RMSF evaluation
The RMSD measures the conformational deviation of the protein construction from the preliminary docked conformation to the ultimate conformation below the dynamic aqueous setting over the simulation time span to make sure the soundness of the expected protein–ligand advanced after ligand binding (Zare et al., 2024; Aier et al., 2016). On this examine, in a simulation time of 100 ns, the CST–18alpha-glycyrrhetinic acid and CST–α-carotene complexes had been discovered to be essentially the most secure with negligible fluctuation, whereas the CST–β-carotene advanced confirmed general stability with little fluctuation at 40 ns The CST–lupeol advanced confirmed preliminary fast fluctuation, however after 25 ns, it additionally achieved stability. The typical deviation discovered for CST, CST-GC, CST–18alpha-glycyrrhetinic acid, CST–α-carotene, CST–β-carotene, and CST-Lupeol was 0.49, 0.76, 0.57, 0.89, 0.92, and 0.67, respectively. Thus, RMSD simulation confirmed that the 4 complexes of CST might preserve general structural stability in the course of the simulation and no vital conformational modifications occurred within the protein construction after ligand binding because the RMSD was maintained all through the simulation both between the RMSD of free CST and the CST-GC advanced or nearer to the RMSD of the CST–GC advanced, which is an efficient indication towards attaining aggressive inhibition (Figures 3IA–D) (Singh and Singh, 2024a).
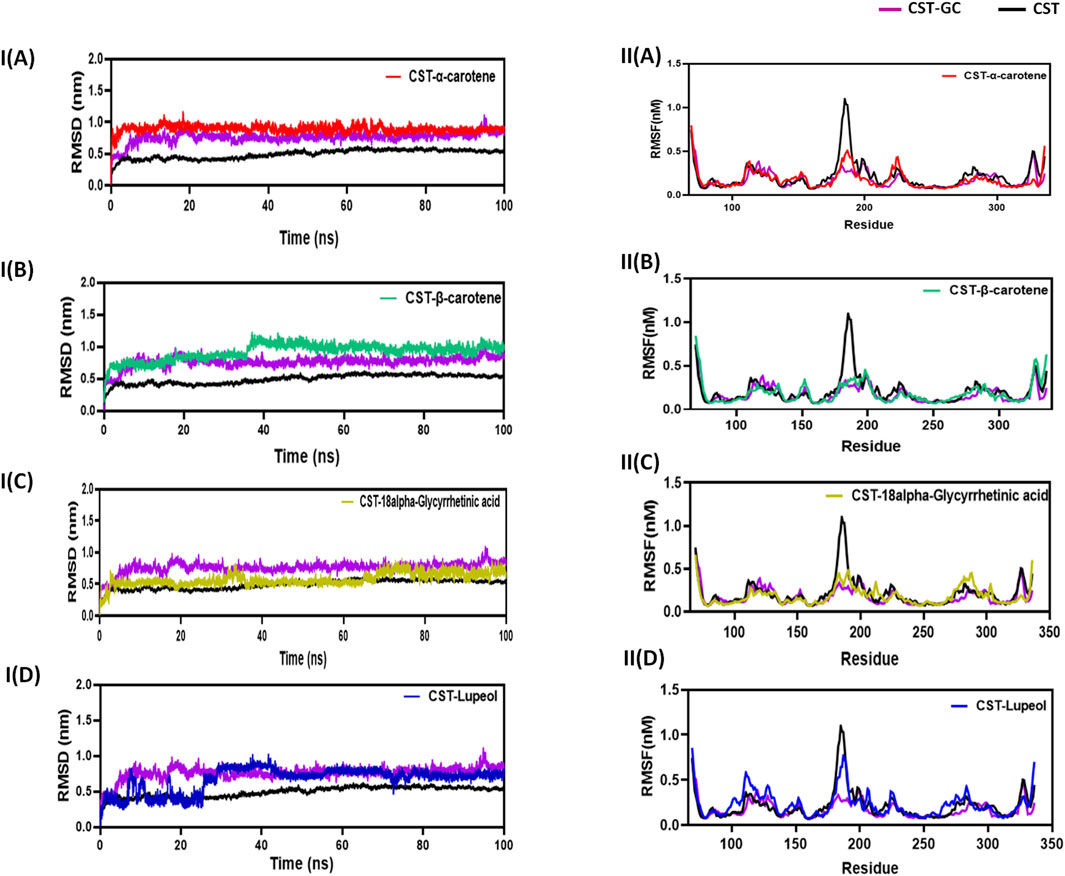
Determine 3. Structural dynamics of CST in advanced with (A) α-carotene, (B) β-carotene, (C) 18alpha-glycyrrhetinic acid, and (D) lupeol. (I) Root imply sq. deviation (RMSD) plot over time, highlighting any deviations within the spatial construction of CST over a time scale of 100 ns from its unique conformation; the plot used a protein spine for contemplating deviation all through simulation. (II) Root imply sq. fluctuation (RMSF) of residues in CST below totally different ligand-bound states throughout a time scale of 100 ns.
Following the protein conformational deviation research with RMSD, protein structural fluctuation on the residue degree was analyzed with RMSF calculation of the protein–ligand advanced (Martínez, 2015; Paul et al., 2022). On this examine, the fluctuations within the residues of the CST protein within the presence of the chosen ligand had been in contrast with the RMSF of the free CST and CST-GC advanced. The typical RMSF of free CST, CST-GC, CST–18alpha-glycyrrhetinic acid, CST–α-carotene, CST–β-carotene, and CST–lupeol was 0.15, 0.17, 0.18, 0.176, 0.18, and 0.23, respectively. As depicted in (Fig 3IIA–D), a wider residual fluctuation was clearly noticed in free CST between the amino acid residues 175 to 190, which represents the loop area within the substrate-binding website, whereas within the CST–GC advanced, residual fluctuation was minimized considerably, as interplay with the ligand might present much less room for fluctuation (Singh and Singh, 2024a). Among the many 4 check complexes, CST–18alpha-glycyrrhetinic acid and CST–β-carotene confirmed comparatively higher stability between amino acid residues 180 and 190, whereas within the CST–lupeol advanced, the fluctuation on this area was near that of the free CST. Total, the CST–18alpha-glycyrrhetinic acid and CST–β-carotene complexes confirmed residual fluctuation near that of the CST–GC advanced, which is once more a great indication of the aggressive inhibition of CST. The CST–lupeol advanced was discovered to be least secure on the residual degree.
3.4.2 Structural compactness with Rg, SASA, and hydrogen bonding evaluation
The following degree of screening was to judge modifications within the dimension or structural compactness of the protein within the presence of ligands in the course of the simulation by measuring Rg of the protein–ligand advanced (Lobanov et al., 2008; Rampogu et al., 2022; Funari et al., 2022). Rg additionally estimates the pliability of the protein below advanced formation by evaluating it with Rg of both the free protein or the substrate-bound protein. The bigger Rg within the free CST indicated lesser compactness and higher flexibility, whereas the smaller Rg within the CST–GC advanced indicated higher compactness and rigidity within the protein construction within the presence of the substrate. As depicted in Figure 4A, an identical pattern was seen with inhibitor binding. Of the 4, the Rg of the CST–β-carotene advanced was most carefully associated to the Rg of the protein–substrate advanced. Within the CST–18alpha-glycyrrhetinic acid advanced, structural compactness of the protein was between that of free CST and the CST-GC advanced, suggesting that the presence of the ligand barely compressed the protein construction. The Rg values of the CST–α-carotene and CST–lupeol complexes had been discovered to be between the Rg of the free CST and the Rg of the CST-GC advanced, additionally suggesting that the protein construction takes on stiffness and compactness within the presence of those ligands (Figure 4A).
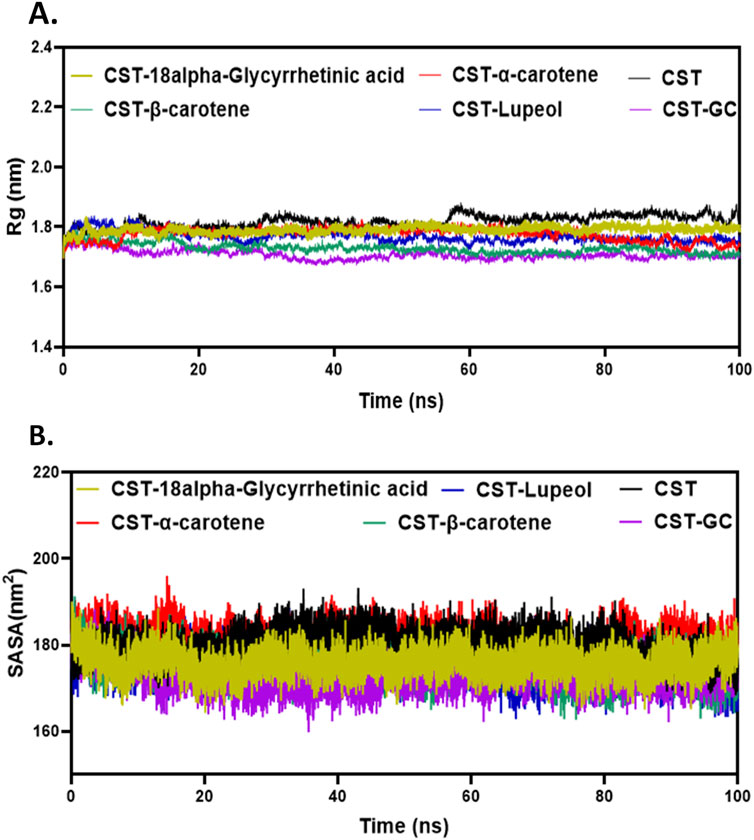
Determine 4. Structural compactness evaluation of CST within the CST–18alpha-glycyrrhetinic acid, CST–α-carotene, CST–β-carotene, and CST–lupeol complexes. Time evolution of (A) Rg values and (B) SASA values in the course of the simulation.
The SASA measures the solvent-accessible floor space of the free protein or protein within the protein–ligand advanced. It mainly calculates the floor space of molecules that’s uncovered to solvent molecules (Alrouji et al., 2023; Durham et al., 2009; Ali et al., 2014; Savojardo et al., 2021). The SASA analyzes how elements of the protein come into contact with the solvent over the simulation time. The typical SASA vary for the chosen compounds was between 170 and 180 nm2, which falls between the vary of the SASA of free CST (180.1 nm2) and that of CST complexed with the substrate (173.07 nm2) (Singh and Singh, 2024a). The typical SASA of the CST–18alpha-glycyrrhetinic acid, CST–α-carotene, CST–β-carotene, and CST–lupeol advanced was 175.83 nm2, 180.89 nm2, 175.38 nm2, and 175.30 nm2, respectively (Figure 4B). Of the 4 hits, the SASA of CST–18alpha-glycyrrhetinic acid, CST–β-carotene, and CST–lupeol falls inside the anticipated vary of the free CST and CST–substrate advanced, suggesting higher compactness of the protein within the presence of those ligands with comparatively lesser undesirable solvent accessibility. The SASA of the CST–α-carotene advanced was near the SASA of free CST, suggesting that ligand binding has a negligible influence on the solvent accessibility of the protein construction. Total, the SASA of the protein–ligand advanced of the chosen compounds confirmed no main modifications within the uncovered protein construction after protein–ligand binding and maintained the pure structural integrity of the protein.
Moreover, intramolecular hydrogen bonding evaluation was important for understanding the soundness of the protein–ligand advanced as ligand binding impacts the general protein intramolecular dynamics (Pace et al., 2014; Hubbard and Haider, 2010). In comparison with the free CST protein, substrate binding to the substrate binding website in CST barely elevated the intramolecular hydrogen bonding, suggesting that when binding to the lively website, the substrate pushed the protein construction inward, facilitating the proximity of atoms and, thus, facilitated extra contacts on the intramolecular degree. Among the many protein–ligand complexes of the chosen compounds, the CST–18alpha-glycyrrhetinic acid, CST–β-carotene, and CST–lupeol complexes confirmed a negligible influence on the intramolecular hydrogen bond dynamics of the CST protein, suggesting the soundness of protein intramolecular dynamics with ligand binding (Figures 5A, C, D). Chance distribution operate (PDF) plots present good consistency of those three complexes. The CST–α-carotene advanced was discovered to be the least secure and most deviated advanced, and the protein construction on this advanced even confirmed negatively extra flexibility than that of the free CST (Figure 5B).
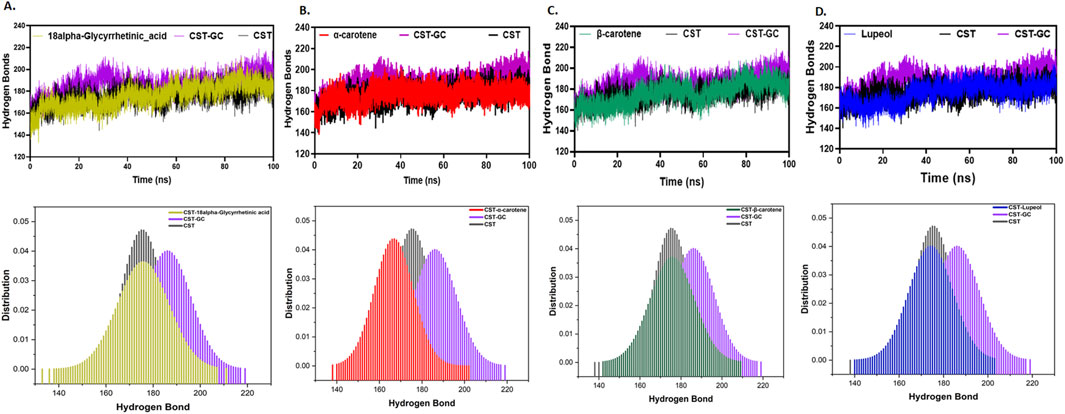
Determine 5. Time evolution of intramolecular hydrogen bond evaluation in CST in (A) CST–18alpha-glycyrrhetinic acid, (B) CST–α-carotene, (C) CST–β-carotene, and (D) CST–lupeol complexes. The decrease panel represents the likelihood distribution operate (PDF) plot of every advanced.
Thus, on the structural compactness degree, the CST–18alpha-glycyrrhetinic acid, CST–β-carotene, and CST–lupeol complexes had been discovered to be comparatively extra secure than the CST–α-carotene advanced. Within the presence of those three compounds, the CST protein underwent the anticipated pure stiffness and compactness phenomenon, as noticed within the case of protein–substrate binding.
3.4.3 Principal part evaluation
The particular catalytic motion of every protein is executed by means of coordinated and collective atomic movement, which determines the soundness of the protein (Zare et al., 2024; Yuan et al., 2024). PCA is broadly used to investigate the atomic-level conformational modifications by means of ligand binding. Mainly, the principal parts are the dominant mode of movement of the system that decide the structural and dynamic properties of the protein–ligand advanced (Zare et al., 2024; Alrouji et al., 2023). On this examine, principal parts for the 4 chosen CST–ligand complexes had been largely decided by the primary two (PC1 and PC2) and the primary three eigenvectors (PC1, PC2, and PC3), which mirrored the general dynamics of the molecular subspace of the CST protein certain with the chosen ligands (Figures 6A, B). With eigenvector 1 spanning between −3.3 and 4.5, eigenvector 2 between −3.07 and three.09, and eigenvector 3 between −3.23 and 4.26, the CST–18alpha-glycyrrhetinic acid advanced confirmed higher compactness among the many 4. Within the CST–β-carotene advanced, eigenvector 1 covers −6.18 to three.87, eigenvector 2 was between −3.3 and a pair of.7, and eigenvector 3 was between −1.7 and three.4. With eigenvector 1 valued between −4.6 and 5.1 nm, eigenvector 2 from −7.3 to 4.1, and eigenvector 3 from −2.0 to 2.7 nm, the CST–α-carotene advanced was discovered to be essentially the most dispersed among the many 4, adopted by the CST–lupeol advanced, of which eigenvector 1 ranged between −4.3 and 5.9, eigenvector 2 from −2.6 to three.8 nm, and eigenvector 3 from −3.6 to 4.6. Within the first two eigenvectors, CST–18alpha-glycyrrhetinic acid, CST–β-carotene, and CST–lupeol complexes fell inside the anticipated vary of free CST and CST complexed with its substrate (GC) and confirmed comparatively much less movement and better stability, whereas the CST–α-carotene advanced was discovered to be essentially the most diverted and fewer secure advanced (Figure 6A). Subsequently, evaluating the outcomes of the primary three principal parts, we discovered that every one 4 complexes primarily fall inside the anticipated vary of the free CST and CST–substrate advanced, with slight diversions noticed within the CST–lupeol advanced (Figure 6B). Thus, in each units of eigenvectors, CST–18alpha-glycyrrhetinic acid and CST–β-carotene demonstrated comparatively higher stability as a result of their principal parts occupied a smaller subspace. Thus, these findings additional strengthen the candidature of 18alpha-glycyrrhetinic acid and β-carotene as inhibitors for CST.
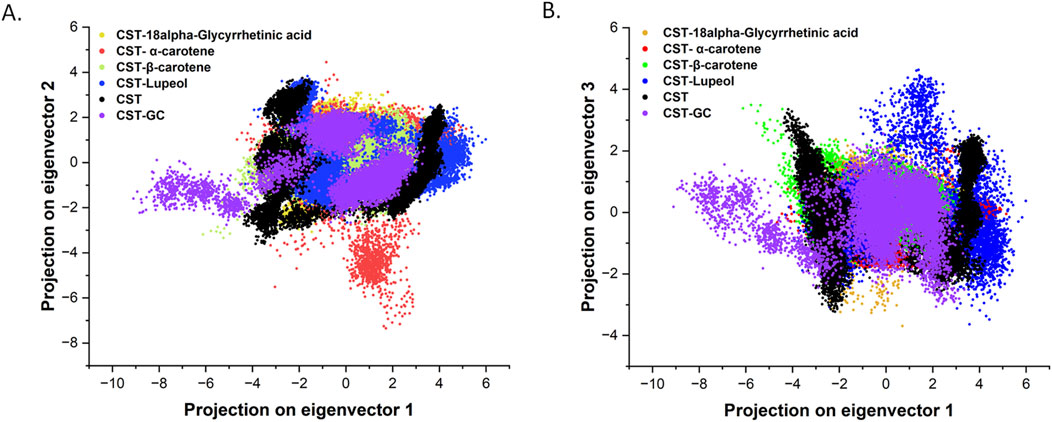
Determine 6. Principal part evaluation of conformational projections of free CST and its complexes substrate (violet), 18alpha-Glycyrrhetinic acid (darkish yellow), α-carotene (purple), β-carotene (mild inexperienced), and Lupeol (blue). (A) Projection of the primary two eigenvectors 1 and a pair of (B) projections of first three eigenvectors and plot between PC1 and PC3.
3.4.4 Free power panorama evaluation
The free power panorama (FEL) evaluation is primarily utilized to grasp the folding sample of proteins and the influence of ligand binding on the construction of the protein (Plattner and Noé, 2015; Lau and Roux, 2007; Khan et al., 2019; Maisuradze et al., 2010). On this examine, the primary two eigenvectors had been used from the principal part evaluation to generate the FEL plot of every protein–ligand advanced based mostly on the dominant and secure conformation throughout a simulation of 100 ns. In distinction to the free CST, which confirmed comparatively elevated conformational house with international minima of 16.60 kJ/mol for attaining a secure construction, the conformational house of CST-GC advanced barely shifted with international minima of 17.10 KJ/mol (Singh and Singh, 2024a). On this examine, within the spectrum of the free power panorama, darkish blue signified essentially the most energetically favored area, and inexperienced depicted the reasonably favored area, the place the conformation of the protein might preserve its stability, whereas yellow represented the comparatively unfavorable area and excessive power state. In accordance with the FEL plot, CST–18alpha-glycyrrhetinic acid, CST–β-carotene, and CST–lupeol complexes confirmed a wider blue zone with a worldwide minima of 16.40 kJ/mol, 17.20 kJ/mol, and 15.10 kJ/mol, respectively (Figures 7A, C, D). The worldwide minima of CST–18alpha-glycyrrhetinic acid and CST–β-carotene complexes had been inside the vary of the worldwide minima of the free CST (16.60 kJ/mol) and CST-GC advanced (17.10 kJ/mol) (Singh and Singh, 2024a), thus exhibiting potential for aggressive inhibition when it comes to power requirement for binding. Among the many 4 hits, the FEL of the CST–α-carotene advanced with a worldwide minima of 19.00 kJ/mol was discovered to have the least secure protein–ligand interplay (Figure 7B). Total, the free power panorama of the protein–ligand complexes supplied a precious perception into the interplay potential of the ligand and the soundness of the protein in its certain type.
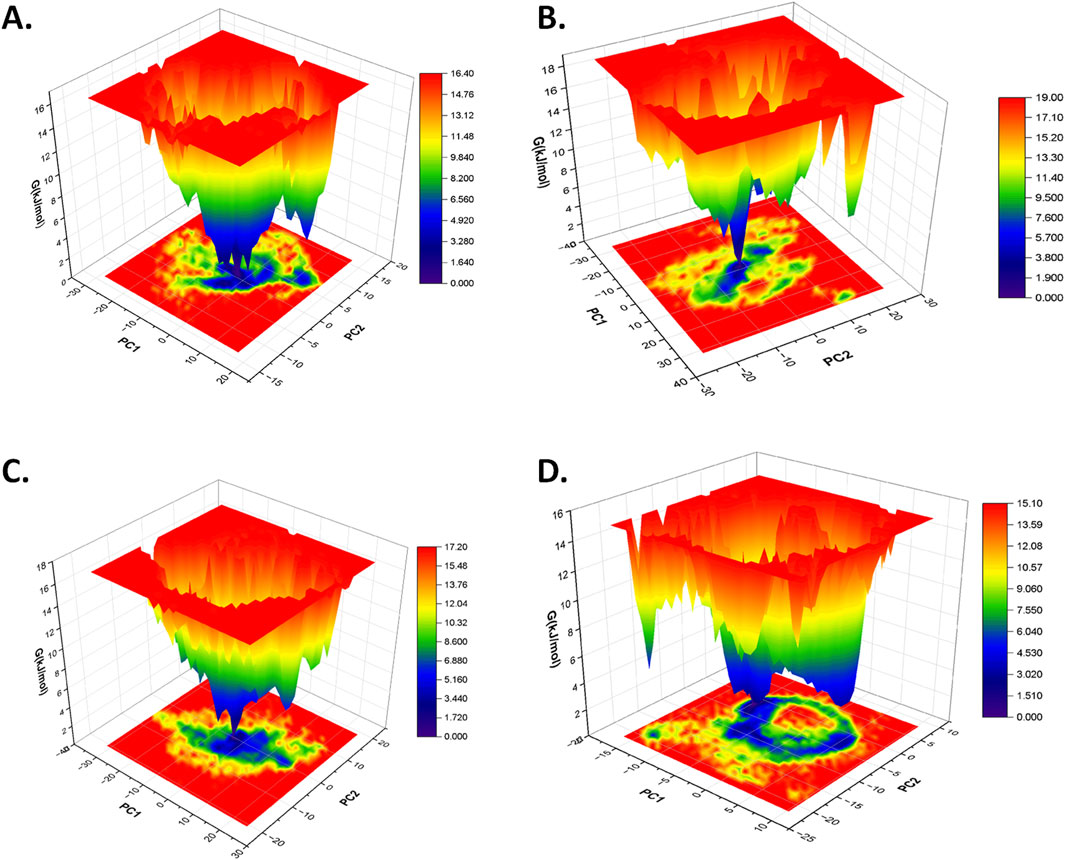
Determine 7. Free power panorama of (A) CST–18alpha-glycyrrhetinic acid, (B) CST–α-carotene, (C) CST–β-carotene, and (D) CST–lupeol complexes.
3.4.5 Dynamic cross-correlation matrix evaluation
Utilizing the MD trajectory knowledge, a DCCM evaluation was carried out to grasp the dominant correlation community of amino acid residues within the CST protein within the presence of ligands (Du et al., 2010; Avti et al., 2022). The dynamic correlation mapping of protein residues within the presence of various ligands indicated the influence of the ligand on the general protein construction conformational stability. For the DCCM examine of the 4 complexes, the MD simulation construction within the final 10 ns was thought-about to acquire an perception into the protein–ligand advanced. Within the 2D matrix of DCCM evaluation, the darkish blue area represents the extremely correlated movement of residues within the constructive path, the white area represents the extremely anti-correlated movement of residues, and the no-color area signifies no correlation within the movement of the residue. Protein residues within the CST–18alpha-glycyrrhetinic acid advanced had been thought-about extremely correlated as a result of they occupied a wider space of darkish blue (Figure 8A). The protein residues within the CST–β-carotene advanced confirmed a comparatively higher residue correlation than that within the CST–α-carotene advanced (Figures 8B, C). The residues within the CST–lupeol advanced had been discovered to have the least correlated residues because the darkish blue area is comparatively sparse among the many 4 protein–ligand complexes, and the big space on this advanced is uncolored, indicating no vital correlation and decreased relation with the goal protein (Figure 8D). Total, the correlated movement of the residue analyzed on this examine confirmed the standard of the protein–ligand advanced. The DCCM evaluation of residues certain to 18alpha-glycyrrhetinic acid revealed the strongest correlation with the protein amongst all 4 complexes, thus indicating a robust protein–ligand interplay. According to different MD trajectory evaluation outcomes, β-carotene additionally confirmed constructive leads to the DCCM evaluation, exhibiting a robust correlation with protein residues.
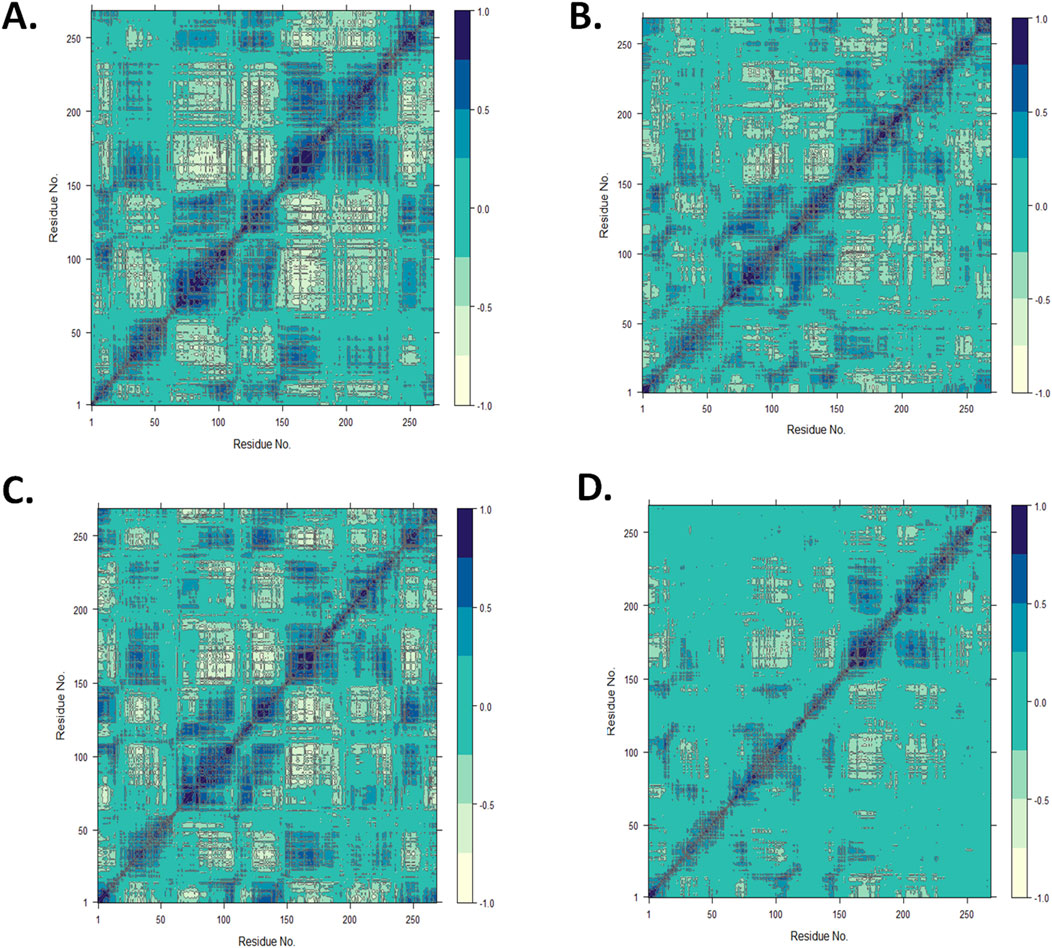
Determine 8. Dynamic cross-correlation map for the protein complexed with (A) 18alpha-glycyrrhetinic acid, (B) α-carotene, (C) β-carotene, and (D) lupeol, utilizing the Bio3D package deal in RStudio.
3.5 Put up-MD protein–ligand interplay pose evaluation
Put up-simulation interplay evaluation was an try to grasp the binding sample of the chosen compounds below a dynamic aqueous setting, the place the protein was free to take its most secure state. Of the 4 hits, 18alpha-glycyrrhetinic acid efficiently retained its interplay with key residues, in addition to its orientation and positioning within the binding pocket of the protein. Within the CST–18alpha-glycyrrhetinic acid advanced, as within the docked conformation, C=O of the carboxylic group shaped a hydrogen bond with Lys85 within the polar website. In the midst of the binding pocket, His84 and His141 flank the compound on each side, and hydrogen bond formation happened with His84, whereas Tyr203 on the fragrant facet interacted with the compound by means of two pi–alkyl bonds (Figure 9A). Moreover, Ser173, as within the docked conformation, retained van der Waals interplay with the C=O group of the third fragrant ring of the compound. Thus, the CST–18alpha-glycyrrhetinic acid advanced confirmed a robust protein–ligand interplay. Throughout simulation, β-carotene additionally largely maintained an identical orientation within the binding pocket, with a slight change within the interplay sample. Lys85 and Tyr176 within the left polar area, His84 and Phe177 within the center, and Tyr203, Phe170, and Arg202 on the proper finish of the binding pocket decided the positioning of β-carotene within the binding pocket (Figure 9C). In distinction to the similarity within the docked conformation of CST–α-carotene and CST–β-carotene complexes, α-carotene was discovered to be the least secure within the binding pocket in the course of the simulation. The isomeric fragrant ring of α-carotene shifted outward from the polar area away from Lys85, which was a key interacting residue within the docked conformation (Figure 9B). The simulation-led structural shift is proven in pre- and post-MD aligned complexes (Figure 10B). As within the docked conformation, the CST–lupeol advanced maintained its place within the binding pocket, occupying principally the appropriate facet of the binding pocket. Resulting from its comparatively small dimension, lupeol left main room for ligand flexibility, which could not be appropriate for aggressive inhibition (Figure 9D). The gap of many of the pi-interactions was comparatively brief in each the CST–18alpha-glycyrrhetinic acid and CST–β-carotene-simulated complexes than their respective docked complexes, suggesting the strongest binding potential of 18alpha-glycyrrhetinic acid and the second strongest binding potential of β-carotene within the protein-binding pocket within the dynamic setting (Tables 3 and 4). Subsequently, based mostly on the interplay sample evaluation and alignment of pre- and post-MD complexes, 18alpha-glycyrrhetinic acid and β-carotene had been discovered to be appropriate candidates for additional reverse pharmacophore evaluation.
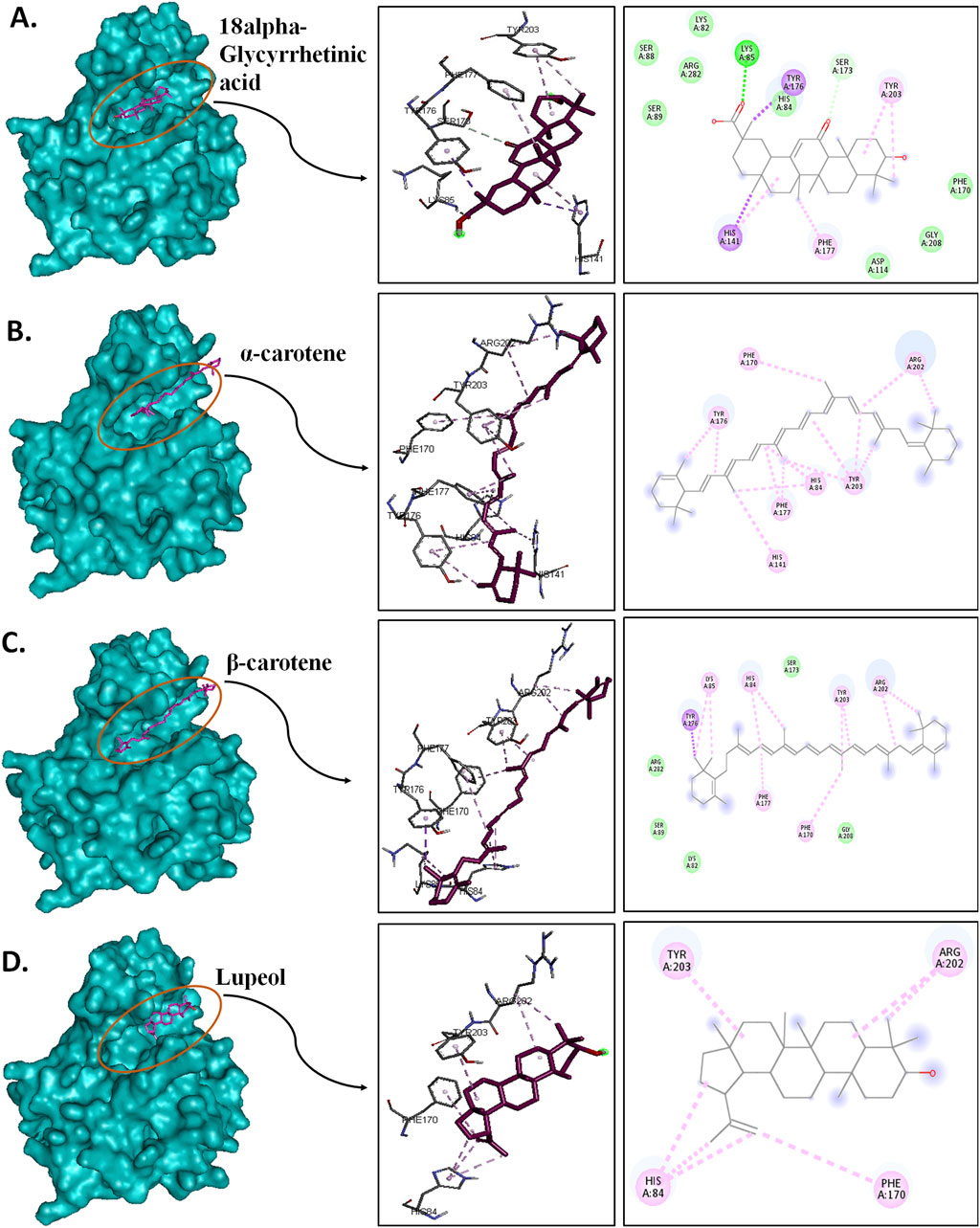
Determine 9. Put up-MD interplay evaluation of protein–ligand complexes with a floor view; 3D and 2D protein–ligand interplay of (A) CST–18alpha-glycyrrhetinic acid, (B) CST–α-carotene, (C) CST–β-carotene, and (D) CST–lupeol. Darkish inexperienced, violet, mild pink, and lightweight inexperienced characterize typical hydrogen bonds, pi–sigma bonds, pi–alkyl bonds, and van der Waals interactions, respectively.
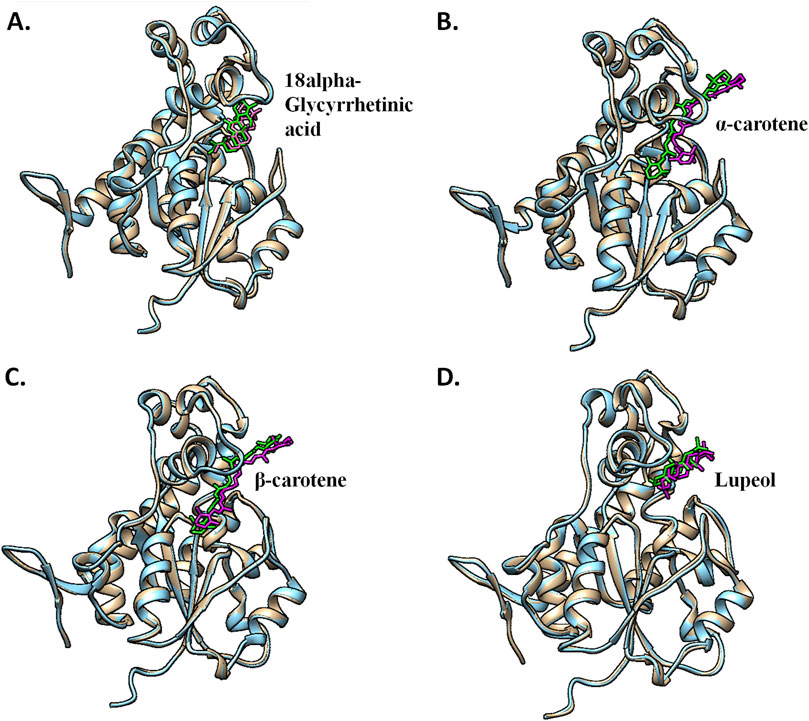
Determine 10. Pre- and post-MD structural alignment of (A) CST–18alpha-glycyrrhetinic acid, (B) CST–α-carotene, (C) CST–β-carotene, and (D) CST–lupeol complexes. Brown signifies the pre-MD CST advanced with inexperienced ligands, and blue depicts the post-simulation protein advanced with magenta ligands.
3.6 Reverse pharmacophore mapping and cross-target identification
Reverse pharmacophore mapping for potential cross-target identification of the CST bioactive compounds (18alpha-glycyrrhetinic acid and β-carotene) was carried out by means of PharmMapper, which in contrast the pharmacophore of the lively compounds with the pharmacophore fashions of 300 proteins deposited within the database (Wang et al., 2017; Liu et al., 2010). Primarily based on their health rating cutoff ≥5.0, the important thing cross-targets had been recognized. Table 5 supplies the small print of the cross-target interactions of 18alpha-glycyrrhetinic acid and β-carotene. The 2 potential targets of 18alpha-glycyrrhetinic acid had been corticosteroid 11-beta-dehydrogenase isozyme 1 and estradiol 17-beta-dehydrogenase 1, and positively, none of those are related to the mind, whereas within the case of β-carotene, potential cross-targets are transthyretin, mobile retinoic acid-binding protein 2, and retinol-binding protein 4 (RBP4), amongst which transthyretin and RBP4 are related to the mind.
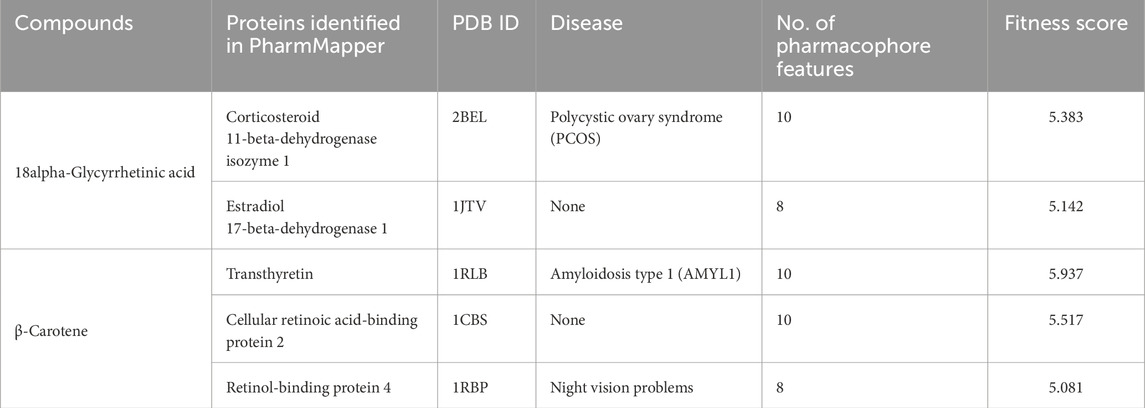
Desk 5. Reverse pharmacophore mapping for figuring out potential targets for the best-performing compounds.
4 Dialogue
Metachromatic leukodystrophy is likely one of the crucial neuropathological situations that come up as a result of accumulation of sulfatides over neurons, which results in the event of white matter abnormalities and, thus, interrupt the correct processing of communication indicators all through the central and peripheral nervous programs. Within the period of bioinformatics development, the choice of creating an oral drug as part of substrate discount remedy is rising as a brand new technique in single-gene issues together with MLD. To counter the main present problem in SRT, i.e., the shortage of the supply of the three-dimensional construction of the goal protein, cerebroside sulfotransferase, our group efficiently developed a 3D homology mannequin of the protein, which was used as a receptor for screening of compounds on this examine. Within the current examine, the substrate-binding website of the protein was used for grid era and protein–ligand interplay examine. For this examine, 4 potential herbs of Medhya Rasayana had been thought-about for screening their phytoconstituents as these herbs are well-regarded as reminiscence enhancers. Initially, based mostly on a binding rating cutoff of ≤−7.5 kcal/mol, the highest 15 compounds had been chosen for performing pharmacokinetics research to get rid of false positives. Together with exhibiting no vital toxicity below ADMET evaluation, the 4 potential compounds—18alpha-glycyrrhetinic acid, α-carotene, β-carotene, and lupeol—confirmed excessive blood–mind barrier permeability, which was thought-about a crucial parameter for the collection of inhibitors because the goal’s bodily location is the mind, and it was essential to pick these compounds that may cross the blood–mind barrier easily.
The interplay evaluation of those 4 compounds in pre- and post-MD confirmed attention-grabbing outcomes, which turned a foundation for the identification of essentially the most potent compounds for CST among the many 4 compounds. Within the pre-MD protein–ligand advanced, the protein was inflexible, and versatile ligands tried to regulate within the lively website. Nonetheless, throughout MD, the protein was subjected to an aqueous setting, which supplied sufficient flexibility to the protein to regulate itself in the absolute best manner to offer most stability to its advanced formation with the ligand. Among the many 4 complexes, 18alpha-glycyrrhetinic acid confirmed a greater interplay with the CST lively website whereas sustaining its place all through the simulation (Figure 10A). The carboxylic acid finish of the compound facilitated the orientation of compounds towards the polar area dominated by Lys85, Lys82, and Tyr176. The center a part of the compound was positioned precisely in the midst of the lively website and flanked by His84 and His141 on each side. Moreover, the oxygen atom on the center of the compound facilitated its tight interplay within the lively website. Phe170, Phe177, and Tyr203 had been key residues within the fragrant website and interacted with the compound through a pi–alkyl interplay. Within the pre-MD interplay examine, α-carotene and β-carotene confirmed an identical interplay sample, with minor variations at their isomeric ends. Nonetheless, after MD, β-carotene maintained its place within the lively website (Figure 10C), whereas α-carotene deviated from its preliminary place towards exterior, as proven in Figure 10B. Primarily based on lively website occupancy standards, lupeol, regardless of sustaining its place within the lively website all through the simulation, was thought to be much less potent because it left sufficient house within the lively website for different ligands to both cross-contaminate the lively website or substitute it if the opposite ligand is obtainable in enough focus (Figure 10D). Subsequently, lupeol was thought-about much less appropriate to fulfill the competitiveness standards.
The in-depth trajectory evaluation of the chosen compounds supplied enough info for understanding the above structural modifications. The RMSD of 4 ligands confirmed stability when it comes to general structural deviation inside the stipulated simulation time. The residual-level fluctuation was sharpened within the RMSF examine, the place the fluctuation was extra pronounced in lupeol, adopted by α-carotene. The CST–18alpha-glycyrrhetinic acid and CST–β-carotene complexes confirmed higher residual stability. Concerning structural compactness parameters together with Rg and SASA, the protein in all 4 complexes largely maintained its structural compactness inside the vary of the free CST and CST–substrate advanced, whereas at intramolecular hydrogen-bond formation-based structural compactness standards, the protein within the CST–α-carotene advanced confirmed least compactness among the many 4 complexes. Moreover, in PCA, the deviation of α-carotene and lupeol was extra pronounced, whereas CST–18alpha-glycyrrhetinic acid and CST–β-carotene had been comparatively higher confined with occupying minimal subspace. An analogous pattern was additionally noticed within the free power panorama evaluation; among the many 4, solely 18alpha-glycyrrhetinic acid and β-carotene confirmed international minima between the vary of the free CST and CST–substrate advanced, whereas α-carotene was far above the vary, and lupeol was discovered far beneath the vary. Within the dynamic cross-correlation map, the CST–lupeol advanced was discovered to be least correlated. Within the correlation matrix, the CST–18alpha-glycyrrhetinic advanced was discovered to be the perfect correlated advanced among the many 4, adopted by the CST–β-carotene advanced. Thus, the general trajectory evaluation represented the CST–α-carotene and CST–lupeol complexes as lesser secure complexes, therefore excluded from additional research.
Reverse pharmacophore mapping was carried out to rule out the chances of cross-contamination with different nervous system proteins. With no potential neurodegenerative illness hyperlink goal, 18alpha-glycyrrhetinic acid may be thought-about essentially the most potent and particular drug candidate in opposition to CST, whereas within the case of β-carotene, transthyretin and RBP4 might act as cross-targets within the mind (Pinheiro et al., 2022; Ishii et al., 2019). Cross-binding of β-carotene to transthyretin might trigger β-amyloid deposition, resulting in neuropathological situations. Within the central nervous system, RBP4 binds and inhibits transthyretin and, thus, regulates the breakdown of amyloid-β, whereas the cross-interaction of β-carotene with RBP4 might cut back the β-amyloid deposition by proscribing the interplay of RBP4 with transthyretin (Pinheiro et al., 2022). To know the advantage of the prevailing discrepancies with the cross-target interactions of β-carotene with different proteins, a robust suggestion is made to additional validate the outcome by means of experimental knowledge.
Thus, the combinatorial high-throughput in silico screening procedures carried out on this examine prompt the general stability of 18alpha-glycyrrhetinic acid and β-carotene contained in the lively website pocket of the CST protein. Though 18alpha-glycyrrhetinic acid was discovered to be the perfect performer all through the screening whereas sustaining the general integrity of the protein conformation throughout advanced formation in a dynamic simulation setting, β-carotene may also be thought-about for additional research because the second-best drug candidate because it additionally confirmed sturdy binding affinity within the binding website pocket of CST. Thus, the appliance of molecular docking, ADMET evaluation, and simulation-led trajectory evaluation supplied an intensive understanding of the general stability of the protein–ligand interplay. Nonetheless, regardless of the a number of ranges of stringent screening performed on this examine, there isn’t a ruling out the potential for failure of the proposed drug candidates on the experimental degree. Subsequently, the efficiency and specificity decided on this examine want additional validation by means of in vitro enzymatic, cell line research and subsequent in vivo validation utilizing animal fashions to finish the preclinical drug discovery validation course of to acquire a potent drug candidate for the medical trial of future marketed medicine. Moreover, since these drug candidates are originated from medicinal vegetation, they will additionally open the door for combinatorial substrate discount remedy the place potent medicine can mix with the hydroalcoholic extract of the entire plant to reduce any type of unintended effects of potent compounds whereas sustaining the efficiency through optimizing the dosage mixture. Thus, the concept is to develop the most secure potential oral medicine for metachromatic leukodystrophy, which is likely one of the most deadly genetic ailments in kids.
5 Conclusion
Cerebroside sulfotransferase is recognized as a lovely goal protein to develop substrate discount remedy as a brand new therapeutic intervention within the discipline of metachromatic leukodystrophy. This examine aimed to determine a potent and selective inhibitor for CST inhibition utilizing a multipronged digital screening strategy utilizing a library of phytoconstituents of common herbs of Medhya Rasayana. The screening of key neuroprotective herbs of Medhya Rasayana is a vital technique to determine potential neuroprotective inhibitors of CST. The phytoconstituents are pure compounds and are extra adaptable to the pure system. Figuring out potent drug candidates will additional strengthen the efficiency of Ayurvedic herbs by making use of the combinatorial technique of therapy whereas minimizing any unfavourable influence of the excessive focus of drug candidates. On this examine, 18alpha-glycyrrhetinic acid was recognized as essentially the most potent and particular inhibitor of CST, whereas β-carotene was discovered to be the second-most potent inhibitor of CST. Thus, the digital screening strategy utilized on this examine is a focused strategy that saved time and useful resource in comparison with the normal strategy of testing random chemical compounds by means of enzyme expertise or making use of a chemical synthesis strategy for testing new drug entities as inhibitors. The MD simulation supplied an perception into the binding sample and orientation of compounds within the lively website by offering data about essentially the most dominant mode of the movement answerable for figuring out the efficiency and selectivity of compounds. This info could be useful for designing new compounds based mostly on the amino acid composition of the lively website pocket. Thus, the findings of the current examine slender down the big pool of datasets and supply a path for additional in vitro and in vivo research.
Knowledge availability assertion
All knowledge generated or analyzed throughout this examine may be discovered on this printed article and its supplementary file, and are additionally obtainable from the corresponding creator upon cheap request.
Writer contributions
NS: conceptualization, knowledge curation, formal evaluation, funding acquisition, investigation, methodology, undertaking administration, sources, software program, supervision, validation, visualization, writing–unique draft, and writing–evaluate and enhancing. AS: supervision, validation, and writing–evaluate and enhancing.
Funding
The creator(s) declare that monetary assist was obtained for the analysis, authorship, and/or publication of this text. Nivedita Singh acknowledges the Institute of Eminence, Banaras Hindu College, Authorities of India, for offering the fellowship and analysis grant for the continued undertaking below the Malaviya Put up-Doctoral Fellowship scheme with Grant ID R/Dev/G/6031/IoE/MPDFs/61698.
Battle of curiosity
The authors declare that the analysis was carried out within the absence of any industrial or monetary relationships that could possibly be construed as a possible battle of curiosity.
Writer’s word
All claims expressed on this article are solely these of the authors and don’t essentially characterize these of their affiliated organizations, or these of the writer, the editors, and the reviewers. Any product that could be evaluated on this article, or declare that could be made by its producer, is just not assured or endorsed by the writer.
Supplementary materials
The Supplementary Materials for this text may be discovered on-line at: https://www.frontiersin.org/articles/10.3389/fmolb.2024.1476482/full#supplementary-material
References
Abrego-Guandique, D. M., Bonet, M. L., Caroleo, M. C., Cannataro, R., Tucci, P., Ribot, J., et al. (2023). The impact of beta-carotene on cognitive operate: a scientific evaluate. Mind Sci. 13 (10), 1468. doi:10.3390/BRAINSCI13101468
Aier, I., Varadwaj, P. Okay., and Raj, U. (2016). Structural insights into conformational stability of each wild-type and mutant EZH2 receptor. Sci. Rep. 6, 34984. doi:10.1038/SREP34984
Ali, S., Hassan, Md., Islam, A., and Ahmad, F. (2014). A evaluate of strategies obtainable to estimate solvent-accessible floor areas of soluble proteins within the folded and unfolded states. Curr. Protein Pept. Sci. 15 (5), 456–476. doi:10.2174/1389203715666140327114232
Alrouji, M., Benjamin, L. S., Alhumaydhi, F. A., Al Abdulmonem, W., Baeesa, S. S., Rehan, M., et al. (2023). Unlocking potential inhibitors for bruton’s tyrosine kinase by means of in-silico drug repurposing methods. Sci. Rep. 13 (1), 17684. doi:10.1038/S41598-023-44956-0
Avti, P., Chauhan, A., Shekhar, N., Prajapat, M., Sarma, P., Kaur, H., et al. (2022). Computational foundation of SARS-CoV 2 most important protease inhibition: an perception from molecular dynamics simulation based mostly findings. J. Biomol. Struct. Dyn. 40 (19), 8894–8904. doi:10.1080/07391102.2021.1922310
Banerjee, P., Eckert, A. O., Schrey, A. Okay., and Preissner, R. (2018). ProTox-II: a webserver for the prediction of toxicity of chemical compounds. Nucleic Acids Res. 46 (Net Server subject), W257–W263. doi:10.1093/NAR/GKY318
Banerjee, S., Baghel, D., Pacheco de Oliveira, A., and Ghosh, A. (2023). β-Carotene, a potent amyloid aggregation inhibitor, promotes disordered aβ fibrillar construction. Int. J. Mol. Sci. 24 (6), 5175. doi:10.3390/ijms24065175
Chang, S. C., Bergamasco, A., Bonnin, M., Bisonó, T. A., and Moride, Y. (2024). A scientific evaluate on the start prevalence of metachromatic leukodystrophy. Orphanet J. Uncommon Dis. 19 (1), 80. doi:10.1186/S13023-024-03044-W
Daina, A., Michielin, O., and Zoete, V. (2017). SwissADME: a free net instrument to judge pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7 (1), 42717–42813. doi:10.1038/srep42717
Du, Q. S., Wang, C. H., Liao, S. M., and Huang, R. B. (2010). Correlation evaluation for protein evolutionary household based mostly on amino acid place mutations and utility in PDZ area. PLoS One 5 (10), e13207. doi:10.1371/JOURNAL.PONE.0013207
Durham, E., Dorr, B., Woetzel, N., Staritzbichler, R., and Meiler, J. (2009). Solvent accessible floor space approximations for fast and correct protein construction prediction. J. Mol. Mannequin 15 (9), 1093–1108. doi:10.1007/S00894-009-0454-9
Eichler, F., Sevin, C., Barth, M., Pang, F., Howie, Okay., Walz, M., et al. (2022). Understanding caregiver descriptions of preliminary indicators and signs to enhance prognosis of metachromatic leukodystrophy. Orphanet J. Uncommon Dis. 17 (1), 370. doi:10.1186/S13023-022-02518-Z
Fernández-Pereira, C., Millán-Tejado, B. S., Gallardo-Gómez, M., Pérez-Márquez, T., Alves-Villar, M., Melcón-Crespo, C., et al. (2021). Therapeutic approaches in lysosomal storage ailments. Biomolecules 11, 1775. doi:10.3390/biom11121775
Forli, S., Huey, R., Pique, M. E., Sanner, M. F., Goodsell, D. S., and Olson, A. J. (2016). Computational protein-ligand docking and digital drug screening with the AutoDock suite. Nat. Protoc. 11 (5), 905–919. doi:10.1038/NPROT.2016.051
Fumagalli, F., Calbi, V., Natali Sora, M. G., Sessa, M., Baldoli, C., Rancoita, P. M. V., et al. (2022). Lentiviral haematopoietic stem-cell gene remedy for early-onset metachromatic leukodystrophy: long-term outcomes from a non-randomised, open-label, section 1/2 trial and expanded entry. Lancet 399 (10322), 372–383. doi:10.1016/S0140-6736(21)02017-1
Funari, R., Bhalla, N., and Gentile, L. (2022). Measuring the radius of gyration and intrinsic flexibility of viral proteins in buffer resolution utilizing small-angle X-ray scattering. ACS Meas. Sci. au 2 (6), 547–552. doi:10.1021/ACSMEASURESCIAU.2C00048
Guterres, H., and Im, W. (2020). Bettering protein-ligand docking outcomes with high-throughput molecular dynamics simulations. J. Chem. Inf. Mannequin 60 (4), 2189–2198. doi:10.1021/ACS.JCIM.0C00057
Hemmerich, J., and Ecker, G. F. (2020). In silico toxicology: from construction–exercise relationships in direction of deep studying and adversarial final result pathways. Wiley Interdiscip. Rev. Comput. Mol. Sci. 10 (4), e1475. doi:10.1002/WCMS.1475
Hira, S., Saleem, U., Anwar, F., Sohail, M. F., Raza, Z., and Ahmad, B. (2019). β-Carotene: a pure compound improves cognitive impairment and oxidative stress in a mouse mannequin of streptozotocin-induced alzheimer’s illness. Biomolecules 9 (9), 441. doi:10.3390/BIOM9090441
Hubbard, R. E., and Haider, M. Okay. (2010). Hydrogen bonds in proteins: position and power. Encycl. Life Sci. doi:10.1002/9780470015902.A0003011.PUB2
Ishii, M., Kamel, H., and Iadecola, C. (2019). Retinol binding protein 4 ranges aren’t altered in preclinical alzheimer’s illness and never related to cognitive decline or incident dementia. J. Alzheimers Dis. 67 (1), 257–263. doi:10.3233/JAD-180682
Istaiti, M., Becker-Cohen, M., Dinur, T., Revel-Vilk, S., and Zimran, A. (2022). Actual-life expertise with oral Eliglustat in sufferers with gaucher illness beforehand handled with enzyme substitute remedy. J. Clin. Med. 11 (21), 6265. doi:10.3390/jcm11216265
Khan, M. T., Khan, A., Rehman, A. U., Wang, Y., Akhtar, Okay., Malik, S. I., et al. (2019). Structural and free power panorama of novel mutations in ribosomal protein S1 (RpsA) related to pyrazinamide resistance. Sci. Rep. 9 (1), 7482–7512. doi:10.1038/s41598-019-44013-9
Khan, S., Farooq, U., and Kurnikova, M. (2016). Exploring protein stability by comparative molecular dynamics simulations of homologous hyperthermophilic, mesophilic, and psychrophilic proteins. J. Chem. Inf. Mannequin 56 (11), 2129–2139. doi:10.1021/ACS.JCIM.6B00305
Komada, N., Fujiwara, T., Yoshizumi, H., Ida, H., and Shimoda, Okay. (2021). A Japanese affected person with gaucher illness handled with the oral drug Eliglustat as substrate decreasing remedy. Case Rep. Gastroenterol. 15 (3), 838–845. doi:10.1159/000519005
Lamichhane, A., and Cabrero, F. R. (2023). Metachromatic leukodystrophy. Frontiers in lysosomal storage ailments (LSD) remedies, 153–162. doi:10.1177/0883073816656401
Lau, A. Y., and Roux, B. (2007). The free power landscapes governing conformational modifications in a glutamate receptor ligand-binding area. Construction 15 (10), 1203–1214. doi:10.1016/J.STR.2007.07.015
Li, W., Guillaume, J., Baqi, Y., Wachsmann, I., Gieselmann, V., Van Calenbergh, S., et al. (2020). Synthesis and structure-activity relationships of cerebroside analogues as substrates of cerebroside sulphotransferase and discovery of a aggressive inhibitor. J. Enzyme Inhib. Med. Chem. 35 (1), 1503–1512. doi:10.1080/14756366.2020.1791841
Liu, X., Ouyang, S., Yu, B., Liu, Y., Huang, Okay., Gong, J., et al. (2010). PharmMapper server: an online server for potential drug goal identification utilizing pharmacophore mapping strategy. Nucleic Acids Res. 38 (Net Server subject), W609–W614. doi:10.1093/NAR/GKQ300
Lobanov, M. Y., Bogatyreva, N. S., and Galzitskaya, O. V. (2008). Radius of gyration as an indicator of protein construction compactness. Mol. Biol. 42 (4), 623–628. doi:10.1134/s0026893308040195
Maisuradze, G. G., Liwo, A., and Scheraga, H. A. (2010). Relation between free power landscapes of proteins and dynamics. J. Chem. Idea Comput. 6 (2), 583–595. doi:10.1021/CT9005745
Martínez, L. (2015). Computerized identification of cellular and inflexible substructures in molecular dynamics simulations and fractional structural fluctuation evaluation. PLoS One 10 (3), e0119264. doi:10.1371/JOURNAL.PONE.0119264
Mistry, P. Okay., Kishnani, P. S., Balwani, M., Charrow, J. M., Hull, J., Weinreb, N. J., et al. (2023). The Two Substrate Discount Therapies for Sort 1 Gaucher Illness Are Not Equal. Touch upon Hughes et al. Switching between Enzyme Substitute Therapies and Substrate Discount Therapies in Sufferers with Gaucher Illness: Knowledge from the Gaucher Consequence Survey (GOS). J. Clin. Med. 2022, 11, 5158. J. Clin. Med. 12 (9), 3269. doi:10.3390/JCM12093269
Miyake, N., Miyake, Okay., Sakai, A., Yamamoto, M., Suzuki, H., and Shimada, T. (2021). Therapy of grownup metachromatic leukodystrophy mannequin mice utilizing intrathecal administration of kind 9 AAV vector encoding arylsulfatase A. Sci. Rep. 11 (1), 20513. doi:10.1038/S41598-021-99979-2
Mohanraj, Okay., Karthikeyan, B. S., Vivek-Ananth, R. P., Chand, R. P. B., Aparna, S. R., Mangalapandi, P., et al. (2018). IMPPAT: a curated database of Indian medicinal vegetation, Phytochemistry and therapeutics. Sci. Rep. 8 (1), 4329–4417. doi:10.1038/s41598-018-22631-z
Narisawa, T., Fukaura, Y., Hasebe, M., Ito, M., Aizawa, R., Murakoshi, M., et al. (1996). Inhibitory results of pure carotenoids, alpha-carotene, beta-carotene, lycopene and lutein, on colonic aberrant crypt foci formation in rats. Most cancers Lett. 107 (1), 137–142. doi:10.1016/0304-3835(96)04354-6
Tempo, C. N., Fu, H., Fryar, Okay. L., Landua, J., Trevino, S. R., Schell, D., et al. (2014). Contribution of hydrogen bonds to protein stability. Protein Sci. 23 (5), 652–661. doi:10.1002/PRO.2449
Paul, S. Okay., Saddam, M., Rahaman, Okay. A., Choi, J. G., Lee, S. S., and Hasan, M. (2022). Molecular modeling, molecular dynamics simulation, and important dynamics evaluation of grancalcin: an upregulated biomarker in experimental autoimmune encephalomyelitis mice. Heliyon 8 (10), e11232. doi:10.1016/J.HELIYON.2022.E11232
Penati, R., Fumagalli, F., Calbi, V., Bernardo, M. E., and Aiuti, A. (2017). Gene remedy for lysosomal storage issues: current advances for metachromatic leukodystrophy and mucopolysaccaridosis I. J. Inherit. Metab. Dis. 40 (4), 543–554. doi:10.1007/S10545-017-0052-4
Pinheiro, F., Pallarès, I., Peccati, F., Sánchez-Morales, A., Varejão, N., Bezerra, F., et al. (2022). Improvement of a extremely potent transthyretin amyloidogenesis inhibitor: design, synthesis, and analysis. J. Med. Chem. 65 (21), 14673–14691. doi:10.1021/acs.jmedchem.2c01195
Pires, D. E. V., Blundell, T. L., and Ascher, D. B. (2015). PkCSM: predicting small-molecule pharmacokinetic and toxicity properties utilizing graph-based signatures. J. Med. Chem. 58 (9), 4066–4072. doi:10.1021/acs.jmedchem.5b00104
Pirolli, D., Righino, B., Camponeschi, C., Ria, F., Di Sante, G., and De Rosa, M. C. (2023). Digital screening and molecular dynamics simulations present perception into repurposing medicine in opposition to SARS-CoV-2 variants spike protein/ACE2 interface. Sci. Rep. 13 (1), 1494. doi:10.1038/S41598-023-28716-8
Plattner, N., and Noé, F. (2015). Protein conformational plasticity and sophisticated ligand-binding kinetics explored by atomistic simulations and markov fashions. Nat. Commun. 6 (1), 7653–7710. doi:10.1038/ncomms8653
Raies, A. B., and Bajic, V. B. (2016). In silico toxicology: computational strategies for the prediction of chemical toxicity. Wiley Interdiscip. Rev. Comput. Mol. Sci. 6 (2), 147–172. doi:10.1002/WCMS.1240
Rampogu, S., Lee, G., Park, J. S., Lee, Okay. W., and Kim, M. O. (2022). Molecular docking and molecular dynamics simulations uncover curcumin analogue as a believable twin inhibitor for SARS-CoV-2. Int. J. Mol. Sci. 23 (3), 1771. doi:10.3390/IJMS23031771
Rashmi, P., Anubha, C., and Gaurav, S. (2017). Idea of Medhya Rasayana in Ayurveda: an summary neelam. Int. J. Res. Ayurveda Pharm. 8 (2). doi:10.7897/2277-4343.08287
Sarokte, A. S., and Rao, M. V. (2013). Results of Medhya Rasayana and yogic practices in enchancment of short-term reminiscence amongst school-going kids. Ayu 34 (4), 383–389. doi:10.4103/0974-8520.127720
Savojardo, C., Manfredi, M., Martelli, P. L., and Casadio, R. (2021). Solvent accessibility of residues present process pathogenic variations in people: from protein buildings to protein sequences. Entrance. Mol. Biosci. 7, 626363. doi:10.3389/fmolb.2020.626363
Sessa, M., Lorioli, L., Fumagalli, F., Acquati, S., Redaelli, D., Baldoli, C., et al. (2016). Lentiviral haemopoietic stem-cell gene remedy in early-onset metachromatic leukodystrophy: an ad-hoc evaluation of a non-randomised, open-label, section 1/2 trial. Lancet 388 (10043), 476–487. doi:10.1016/S0140-6736(16)30374-9
Shaimardanova, A. A., Chulpanova, D. S., Solovyeva, V. V., Mullagulova, A. I., Kitaeva, Okay. V., Allegrucci, C., et al. (2020). Metachromatic leukodystrophy: prognosis, modeling, and therapy approaches. Entrance. Med. (Lausanne) 7, 576221. doi:10.3389/FMED.2020.576221
Singh, N., Malik, A. H., Iyer, P. Okay., and Patra, S. (2021). Diversifying the xanthine scaffold for potential phosphodiesterase 9A inhibitors: synthesis and validation. Med. Chem. Res. 30 (6), 1199–1219. doi:10.1007/s00044-021-02722-9
Singh, N., Patra, S., and Patra, S. (2018). Identification of xanthine derivatives as inhibitors of phosphodiesterase 9A by means of in silico and organic research. Comb. Chem. Excessive. Throughput Display screen 21 (7), 476–486. doi:10.2174/1386207321666180821100713
Singh, N., Saravanan, P., Thakur, M. S., and Patra, S. (2017). Improvement of xanthine based mostly inhibitors concentrating on phosphodiesterase 9A. Lett. Drug Des. Discov. 14 (10), 1122–1137. doi:10.2174/1570180813666161102125423
Singh, N., and Singh, A. Okay. (2024a). In silico structural modeling and binding website evaluation of cerebroside sulfotransferase (CST): a therapeutic goal for creating substrate discount remedy for metachromatic leukodystrophy. ACS Omega 9, 10748–10768. doi:10.1021/acsomega.3c09462
Singh, N., and Singh, A. Okay. (2024b). A complete evaluate on structural and therapeutical perception of cerebroside sulfotransferase (CST) – an essential goal for improvement of substrate discount remedy in opposition to metachromatic leukodystrophy. Int. J. Biol. Macromol. 258, 128780. doi:10.1016/J.IJBIOMAC.2023.128780
Stielow, M., Witczyńska, A., Kubryń, N., Fijałkowski, Ł., Nowaczyk, J., and Nowaczyk, A. (2023). The bioavailability of drugs-the present state of data. Molecules 28 (24), 8038. doi:10.3390/MOLECULES28248038
Vivek-Ananth, R. P., Mohanraj, Okay., Sahoo, A. Okay., and Samal, A. (2023). IMPPAT 2.0: an enhanced and expanded phytochemical atlas of Indian medicinal vegetation. ACS Omega 8 (9), 8827–8845. doi:10.1021/acsomega.3c00156
Wang, X., Shen, Y., Wang, S., Li, S., Zhang, W., Liu, X., et al. (2017). PharmMapper 2017 replace: an online server for potential drug goal identification with a complete goal pharmacophore database. Nucleic Acids Res. 45 (W1), W356–W360. doi:10.1093/NAR/GKX374
Yaghootfam, A., Sorkalla, T., Häberlein, H., Gieselmann, V., Kappler, J., and Eckhardt, M. (2007). Cerebroside sulfotransferase varieties homodimers in dwelling cells. Biochemistry 46 (32), 9260–9269. doi:10.1021/BI700014Q
Yang, H., Solar, L., Li, W., Liu, G., and Tang, Y. (2018). Corrigendum: in silico prediction of chemical toxicity for drug design utilizing machine studying strategies and structural alerts. Entrance. Chem. 6, 129. doi:10.3389/fchem.2018.00129
Yasuo, N., and Sekijima, M. (2019). Improved methodology of structure-based digital screening through interaction-energy-based studying. J. Chem. Inf. Mannequin 59 (3), 1050–1061. doi:10.1021/ACS.JCIM.8B00673
Yuan, F., Li, T., Xu, X., Chen, T., and Cao, Z. (2024). Identification of novel PI3Kα inhibitor in opposition to gastric most cancers: QSAR-molecular docking-and molecular dynamics simulation-based evaluation. Appl. Biochem. Biotechnol. doi:10.1007/S12010-024-04898-3
Zare, F., Ataollahi, E., Mardaneh, P., Sakhteman, A., Keshavarz, V., Solhjoo, A., et al. (2024). A mixture of digital screening, molecular dynamics simulation, MM/PBSA, ADMET, and DFT calculations to determine a possible DPP4 inhibitor. Sci. Rep. 14 (1), 7749. doi:10.1038/S41598-024-58485-X
Zhao, M., Ma, J., Li, M., Zhang, Y., Jiang, B., Zhao, X., et al. (2021). Cytochrome P450 enzymes and drug metabolism in people. Int. J. Mol. Sci. 22 (23), 12808. doi:10.3390/IJMS222312808