Protein coabundance scores genome-wide protein associations
We began by accumulating protein abundance knowledge from proteomics research of cohorts of contributors with most cancers. In whole, we compiled a dataset of fifty research throughout 14 human tissues, encompassing 5,726 samples of tumors and a couple of,085 samples of adjoining wholesome tissue26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75 (Fig. 1a and Supplementary Desk 1). We additional included the mRNA expression knowledge paired to the proteomics for two,930 of the tumor and 722 of the wholesome samples. Following earlier research, we used the truth that protein advanced members are strongly transcriptionally and post-transcriptionally coregulated to compute chances of protein–protein associations from the abundance knowledge5,17,20 (Fig. 1b, Methods and Supplementary Fig. 1). In brief, we preprocessed the abundance knowledge to acquire a log-transformed and median-normalized abundance throughout contributors. For every research, we then computed a coabundance estimate of a protein pair because the Pearson correlation when each proteins had been quantified in at the least 30 samples (Supplementary Fig. 2). Lastly, with pairs of subunits for curated steady protein complexes as ground-truth positives (CORUM76), we used a logistic mannequin for every research to transform the coabundance estimates to chances of protein–protein associations (Supplementary Figs. 3–5).
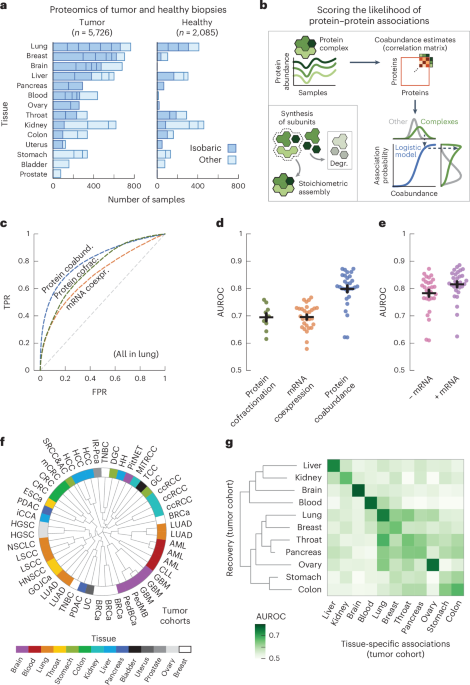
a, Variety of tumor and wholesome samples per tissue. Bar sections point out particular person research, utilizing multiplexed proteomics with isobaric labeling (darkish blue) or different strategies (mild blue). b, Schematic illustration of workflow. Subunits of protein complexes happen in fastened stoichiometries. Protein coabundance is estimated by means of correlation of protein abundance profiles and transformed to chances by means of a logistic mannequin utilizing interactions between subunits of protein complexes (CORUM) as positives. Degr., degradation. c, ROC curves for affiliation chances in lung tissue derived from protein coabundance (coabund.; blue), mRNA coexpression (coexpr.; orange) and protein cofractionation (cofrac.; inexperienced). The grey dashed line exhibits the efficiency of a random classifier. FPR, false-positive charge; TPR, true-positive charge. d, AUC values for affiliation chances as illustrated in c. Proven are research that quantified each protein coabundance (blue; n = 29) and mRNA coexpression (orange; n = 29) or protein cofractionation (inexperienced; n = 10). e, AUC values for affiliation chances derived from protein coabundance mixed with mRNA coexpression by means of a linear mannequin (purple) and protein coabundance after regressing gene expression out of the protein abundance (pink). Proven are the identical research as in d. In d,e, every dot represents one research with paired transcriptomics and proteomics knowledge. Protein pairs had been filtered for having affiliation chances from each modalities. Error bars present the imply and s.e.m. In c–e, negatives are all quantified protein pairs not reported as advanced members. f, Clustering of the n = 48 cohorts utilizing affiliation chances of protein pairs with essentially the most variable associations (CV above the median). The radial dendrogram exhibits complete-linkage clustering with the Pearson correlation distance. Cohorts are labeled in keeping with the kind of most cancers; colours characterize the completely different human tissues. Leaf-joint distances had been shortened. g, Warmth map of AUCs for recovering tissue-specific associations with cohorts that had been withheld when predicting these associations. Every sq. represents the typical AUC for all cohorts of a given tissue. Tissues had been clustered by means of complete-linkage clustering with the Manhattan distance.
To check the flexibility of the affiliation chances to get well recognized advanced members, we computed receiver working attribute (ROC) curves for chances derived from protein coabundance, mRNA coexpression and protein cofractionation6,7,77 (Fig. 1c). We discovered that protein coabundance (space underneath the curve (AUC) = 0.80 ± 0.01 (imply ± s.e.m.)) outperformed protein cofractionation (AUC = 0.69 ± 0.01) and mRNA coexpression (AUC = 0.70 ± 0.01) knowledge for recovering recognized interactions (Fig. 1d and Methods). As well as, the mixture of mRNA and protein abundance knowledge didn’t considerably enhance the restoration of recognized advanced members (Fig. 1e; AUC = 0.82 ± 0.01, P = 0.15, in keeping with a one-sided Welch’s t-test). Due to this fact, with roughly half of all cohorts having paired mRNA expression knowledge accessible, we selected to solely use protein coabundance for computing affiliation chances. Moreover, we discovered related AUCs when regressing gene expression out of the protein abundance earlier than computing protein coabundance estimates (AUC = 0.78 ± 0.01, P = 0.18), suggesting that post-transcriptional processes however not regulation of gene expression drive a lot of the predictive energy for protein associations.
Having established that the affiliation chances derived from protein coabundance knowledge get well recognized interactions of protein advanced members, we sought to check whether or not replicate research of the identical tissue yielded affiliation chances that had been consultant for every tissue. As a place to begin, we used the gene expression knowledge to ascertain that the affiliation chances weren’t pushed by cell-type composition78 (Supplementary Fig. 6). Subsequent, utilizing the 1,115,405 affiliation chances that had been quantified for all research, we discovered that the replicate cohorts from the identical tissue typically clustered collectively (Fig. 1f; for instance, blood, mind, liver and lung). Subsequent, we chosen the associations that had been tissue particular, that’s associations whose common likelihood exceeded the ninety fifth percentile for a given tissue (0.68 ± 0.01 throughout tissues) and whose common likelihood remained under 0.5 throughout all different tissues. By a hold-one-out methodology, we discovered that the tissue-specific associations had been primarily recovered by cohorts of the identical tissue of origin (AUC = 0.71 ± 0.01) in comparison with cohorts from completely different tissues (AUC = 0.56 ± 0.00, P < 0.05 for all tissues, in keeping with a one-sided Welch’s t-test) (Fig. 1g, Methods and Supplementary Fig. 7). Collectively, these observations recommend that the tissue of origin is a serious driver of variations between cohorts.
An atlas of protein associations in human tissues
With the replicate cohorts representing the tissue of origin, we aggregated the affiliation chances from cohorts of the identical tissue into single affiliation scores for 11 human tissues (Fig. 2a and Methods). Aggregating the replicate cohorts was advantageous, as all however one of many particular person cohorts had been outperformed by the tissue-level scores for recovering recognized protein interactions (P = 1.3 × 10−9, in keeping with a one-sided Welch’s t-test). Furthermore, the tumor-derived scores outperformed the healthy-tissue-derived scores for all tissues (Fig. 2b; AUC = 0.87 ± 0.01 and 0.82 ± 0.01, respectively, P = 8.3 × 10−5, in keeping with a one-sided Welch’s t-test). Along with the biopsy, the place the genetic heterogeneity of tumors elevated variation between samples (Supplementary Fig. 8), we discovered a number of different elements affecting the restoration of recognized interactions, such because the accessible variety of cohorts per tissue, the variety of samples per cohort, the tissue of origin and the MS methodology (Supplementary Figs. 2 and 9). The healthy-tissue-derived and tumor-derived scores originated from separate dissections of the identical tissues and contributors and will, thus, function impartial replicates. Analogous to the cohorts, we computed tissue-specific associations from the healthy-tissue-derived scores, which we then recovered with the tumor-derived scores (Fig. 2c). For all tissues, we discovered that the tumor-derived scores primarily recovered the tissue-specific associations of the identical wholesome tissue (AUC = 0.74 ± 0.02) in comparison with the opposite wholesome tissues (AUC = 0.53 ± 0.01, P = 5.9 × 10−5, in keeping with a one-sided Welch’s t-test). These analyses present that the coabundance-derived tissue-level affiliation scores get well recognized protein interactions and are reproducible and consultant of the tissue of origin (Supplementary Fig. 10).
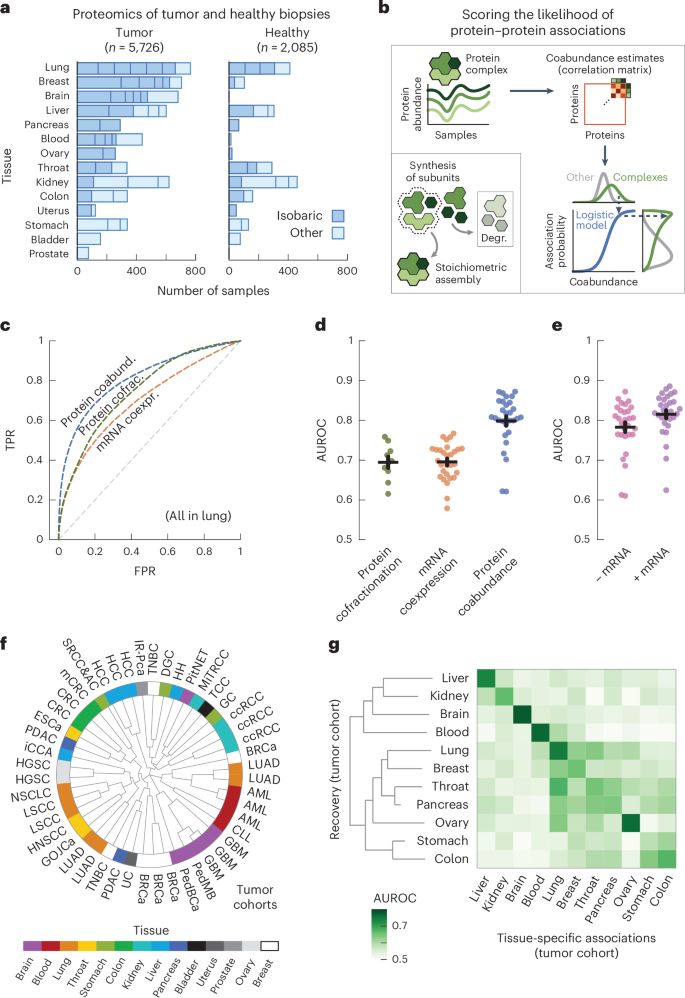
a, Schematic for aggregating replicate cohorts right into a single affiliation rating for a tissue. b, AUC values for the affiliation scores derived from wholesome samples (inexperienced; n = 6) and tumor samples (blue; n = 11), utilizing interactions between subunits of protein complexes (CORUM) as positives. Affiliation scores had been filtered for protein pairs having chances in all cohorts of a tissue. c, Warmth map of AUCs for utilizing tumor-derived affiliation scores to get well tissue-specific associations outlined by the affiliation scores from wholesome tissues. Affiliation scores solely embody cohorts that had each wholesome and tumor samples. Tissues had been clustered by means of complete-linkage clustering with the Manhattan distance. d, Atlas of protein associations in n = 11 human tissues. The radial diagram exhibits, for every tissue, the numbers of protein pairs that had been quantified (grey), are prone to work together (mild inexperienced; affiliation rating > 0.5) or had been confidently quantified (darkish inexperienced; affiliation rating > 0.8). The bar graph exhibits the variety of associations that had been quantified within the given variety of tissues. e, Chance of associations of a tissue to probably be in a healthy-tissue-derived replicate (orange; n = 12) or between pairs of tissues (inexperienced; n = 110) as a operate of threshold affiliation rating. Scores solely embody protein pairs quantified for each tissues or replicates. Proven is the median likelihood throughout pairs of replicates or tissues. The shaded space exhibits the interquartile vary. f, Probably associations shared between pairs of tissues as quantified by the Jaccard index (grey dots), in comparison with shared associations restricted to advanced members (CORUM), bodily associations (STRING scores > 400), organic pathways (Reactome) and signaling (SIGNOR) (purple dots) or associations detected by means of yeast two-hybrid (HuRI) or AP (BioPlex) experiments (blue dots). Every dot represents a pair of tissues. Error bars present the imply and s.e.m. (n = 55).
We outlined a protein affiliation atlas with affiliation scores for all quantified protein pairs by averaging the affiliation chances over the cohorts of every tissue. The ensuing affiliation atlas scores the affiliation probability for 116 million protein pairs throughout 11 human tissues (Fig. 2d). On common, every tissue incorporates affiliation scores for 56 ± 6.2 million protein pairs, of which 10 ± 1.0 million are prone to be related (rating > 0.5, common accuracy = 0.81 over all tissues, recall = 0.73 and diagnostic odds ratio = 13.0) and 0.49 ± 0.08 million are ‘assured’ associations (rating > 0.8, common accuracy = 0.99 throughout tissues, recall = 0.21 and diagnostic odds ratio = 31.9) (Supplementary Fig. 11). These protein associations tended to be probably and assured in only some tissues, with 99,103 protein pairs having probably associations in all tissues (Fig. 2d and Supplementary Fig. 12).
Variations between tissues not pushed by gene expression
One of many well-known drivers of variations in protein interactions between tissues is gene expression; proteins can work together provided that their gene is expressed in a tissue. Certainly, the proteins that had been quantified in a given tissue had been typically enriched for genes with elevated expression for that very same tissue however not the opposite tissues (Supplementary Fig. 13; P = 1.3 × 10−6, in keeping with a one-sided Mann–Whitney U-test). Nevertheless, solely as much as 7% of variations in (probably) associations between tissues will be defined by variations in gene expression and solely by means of the shortage of detection (Supplementary Fig. 14). These observations reveal that the probably associations for every tissue mirror however will not be outlined by variations in gene expression, additional supporting our earlier statement that protein coabundance is primarily pushed by post-transcriptional processes.
Having established that affiliation scores typically reproduce nicely and that variations between tissues will not be pushed by gene expression, we sought to measure the share of tissue-specific associations. To take action, we used a threshold affiliation rating to quantify the share of a tissue’s associations that had been probably (rating > 0.5) for the replicate (Fig. 2e, orange curve, evaluating healthy-tissue-derived and tumor-derived replicates). As anticipated, we discovered that the share of probably associations will increase with the brink rating, with 46.3% of probably associations and 90.2% of assured associations (rating > 0.8) additionally being probably for the replicate tissue. Equally, we discovered that these percentages decreased to 32.9% and 54.6%, respectively, when evaluating associations between pairs of tissues from the affiliation atlas (Fig. 2e, inexperienced curve). Lastly, relying on threshold scores, we discovered between 18.8% and 34.0% (interquartile vary) of probably associations to be tissue particular between pairs of tissues, given the distinction in chances between the curves for replicates and tissues. Due to this fact, with as much as 7% of probably associations not quantified in different tissues due to gene expression (Supplementary Fig. 14), we estimated over 25.8% (18.8% + 7%) of probably associations to be tissue particular.
Tissues get well tissue-specific mobile elements
We sought to characterize the probably associations that had been shared between tissues. In comparison with all probably associations (common Jaccard index = 0.19), we discovered that the similarity between pairs of tissues elevated as we restricted the probably associations to interactions recognized by means of high-throughput screens equivalent to yeast two-hybrid (Jaccard index = 0.30; HuRI)3 or AP (Jaccard index = 0.41; BioPlex)4 experiments (Fig. 2f). Likewise, the similarity between pairs of tissues elevated when limiting probably associations to recognized interactions reported for signaling (Jaccard index = 0.32; SIGNOR)79, organic pathways (Jaccard index = 0.48; Reactome)80, bodily associations (Jaccard index = 0.56; STRING)81 or human protein complexes (Jaccard index = 0.74; CORUM)76. Lastly, we discovered that the quantified variations and similarities between tissues weren’t delicate to the selection of rating cutoffs (Supplementary Fig. 15). Thus, recognized protein interactions are sometimes shared by the tissues in our affiliation atlas, with signaling interactions being much less generally recovered between tissues than steady protein complexes. These observations mirror the divergence between tissues for various kinds of interactions and might also mirror variations in accuracy for recovering associations for steady protein complexes in comparison with spatiotemporal interactions which can be dynamic.
Effectively-characterized protein complexes had been typically preserved throughout tissues, turning into extra variable because the complex-averaged affiliation scores decreased (ρ = −0.77, P = 6.2 × 10−125) (Supplementary Fig. 16). As seen in different proteomics datasets82, extra variable complexes are sometimes concerned signaling and regulation (for instance, tumor necrosis issue and emerin), whereas extra steady complexes are concerned central mobile buildings (for instance, ribosomes and the respiratory chain) (Supplementary Fig. 16). Whereas protein complexes assorted little between tissues, we discovered that associations assorted strongly for tissue-specific mobile elements, for instance, for the mind (synapse-related elements), throat (structural elements of muscle fiber), lung (motile cilia) and liver (peroxisomes) (Supplementary Fig. 17). This implies that tissue-specific and cell-type-specific mobile elements are an vital driver of tissue-specific protein associations which can be impartial of straightforward expression variations.
Affiliation atlas reveals cell-type-specific associations
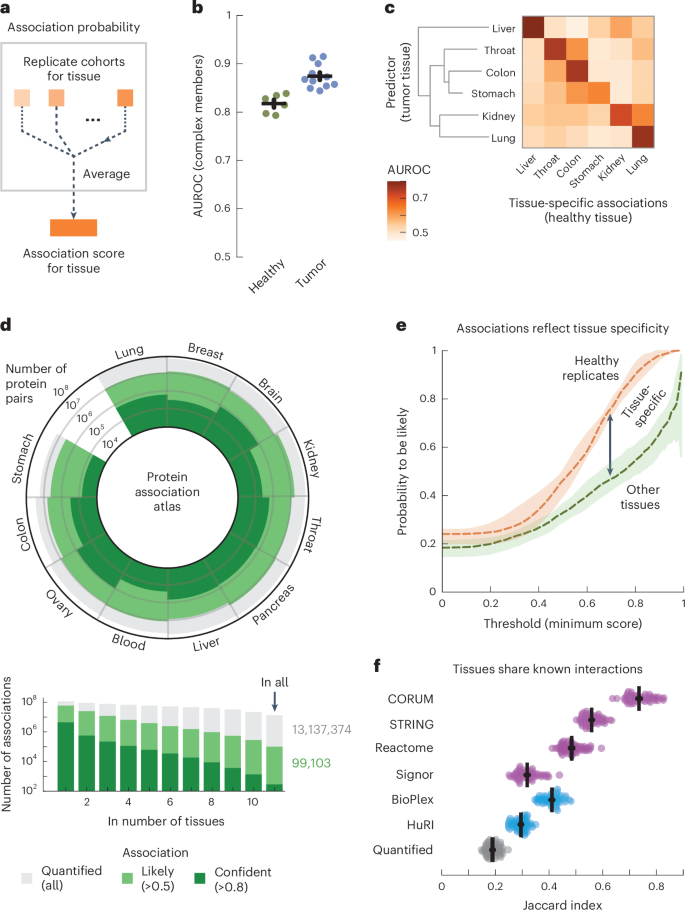
To discover cell-type-specific associations in our affiliation atlas, we took the AP2 adaptor advanced as a widely known instance. The AP2 advanced has neuron-specific capabilities along with capabilities which can be common to all cells83. Certainly, the subunits of the AP2 advanced had been coabundant in all tissues (common affiliation rating between subunits = 0.80). We discovered 91 proteins that had affiliation scores with all AP2 subunits in all tissues and had been recognized to affiliate with AP2 (STRING rating > 400). Amongst these, the 51 synaptic proteins (SynGO84) had larger affiliation scores with the AP2 advanced within the mind (common rating = 0.54) in comparison with the opposite tissues (common rating = 0.48 ± 0.00, P = 6.7 × 10−6, in keeping with a one-sided Mann–Whitney U-test). Conversely, the nonsynaptic interactors had decrease affiliation scores with the AP2 advanced within the mind (common rating = 0.33) in comparison with the opposite tissues (common rating = 0.43 ± 0.00, P = 1.1 × 10−21, in keeping with a one-sided Mann–Whitney U-test) (Fig. 3a). We explored additional examples by specializing in cell-type-specific associations within the context of illness. We discovered that proteins of hemoglobin are associated to anemia and have probably associations with anemia proteins however solely within the blood (Fig. 3b and Methods). Likewise, we discovered that subunits of chylomicron, which transports dairy lipids from the intestines, comprise and have probably associations with proteins associated to Crohn’s illness however solely within the colon85,86. Lastly, we discovered that subunits of fibrinogen, synthesized within the liver, comprise and have liver-only probably associations with proteins associated to liver illness87,88. For the opposite tissues, we may discover many examples of tissue-specific and cell-type-specific associations for protein complexes, mobile elements and problems equivalent to diabetes and bronchial asthma (Supplementary Fig. 18). These examples reveal that our affiliation atlas can be utilized to check tissue-specific capabilities of protein complexes and context-dependent associations for illness genes.
a, Affiliation scores between AP2 subunits and recognized AP2 interactors (STRING scores > 400) which can be synaptic proteins (NECAP1 and BIN1; SynGO) or not (DAB2 and NECAP2). Warmth maps present affiliation scores within the mind and averaged affiliation scores for the opposite tissues. b, Associations of hemoglobin (GO:0005833) to anemia, chylomicron (GO:0042627) to Crohn’s illness and fibrinogen (GO:0005577) to liver illness. Proteins (nodes) are advanced members (grey) and illness genes (black edge). Associations are probably in all tissues (skinny grey strains) or probably in a single tissue and unlikely in all others (thick coloured strains). Thick grey strains are associations with prior proof (STRING scores > 400). Illness genes outlined by means of OTAR (Methods). c, Schematic of strategy. Relationships are scored by aggregating the affiliation scores between all pairs of proteins from disjoint units. d, Relationship scores of mobile elements (mild grey; GO), GWAS traits (darkish blue; OTAR L2G ≥ 0.5) and between traits and elements (mild blue). Every dot represents the connection between two units, indicating the typical and CV of relationship scores relative to the tissue median. Inexperienced dots present relations of the ribosome and spliceosome (rating > 1.75; inexperienced field); purple dots present relations of synaptic elements (CV > 0.4; purple field). Comp., part. e, Dendrograms of the 15 most brain-specific GWAS traits (left) and the 15 GO mobile elements having essentially the most brain-specific relationship with OCD (L2G ≥ 0.5; proper). Dendrograms had been constructed with complete-linkage clustering utilizing the Manhattan distance on the connection scores between traits (left) or between mobile elements (proper). Warmth maps present genes overlapping between mobile elements and OCD (orange; Jaccard index) or the enrichment of nonoverlapping genes from mobile elements with drug targets, genes related to OCD in mice or genes much less confidently linked to OCD by means of GWAS (purple–inexperienced; conditional log2 odds ratios of one-sided Fisher actual take a look at; dots present BH-adjusted P values < 0.05; Methods). SV, synaptic vesicle; CCV, clathrin-coated vesicle; m., membrane; SC, Schaffer collateral; ASD, autism spectrum dysfunction.
Tissue-specific relations of traits and mobile elements
We sought to generalize these examples of context-specific associations by systematically mapping the relationships amongst traits and multiprotein buildings equivalent to complexes or mobile elements. As units of proteins, we outlined mobile elements by Gene Ontology (GO) and outlined human traits on the idea of the genome-wide affiliation research (GWAS) Open Targets (OTAR) locus-to-gene (L2G) rating (≥0.5)89,90,91. We then scored the connection between units of proteins with the median affiliation rating of all doable protein pairs between the units (Fig. 3c, schematic, omitting the intersection between gene units). In whole, we scored the relationships between traits (107,306 pairs), between traits and mobile elements (240,967 pairs) and between elements (134,002 pairs) throughout all tissues (Fig. 3d and Supplementary Tables 2–4). The connection scores that had been excessive in all tissues had been primarily of core mobile elements such because the ribosome and spliceosome (72% of relationships with relative common rating > 1.75), whereas the connection scores that assorted most throughout tissues typically concerned tissue-specific buildings equivalent to synaptic elements (61% of relationships with a coefficient of variation (CV) > 0.4). These observations recommend that the connection scores recapitulate the relatedness of protein units in a tissue-specific method, significantly for the mind.
Relationship scores for prioritizing illness genes
Moreover, we unbiasedly scored the tissue specificity of every protein set because the median affiliation rating between all pairs of its proteins (Methods and Supplementary Tables 5 and 6). Utilizing these scores, we then chosen the 15 traits most particular to the mind, 13 of which had been certainly associated to the mind (Supplementary Desk 7). Clustering these traits utilizing the trait–trait relationship scores from the mind revealed a hierarchical group of traits with co-occurring situations equivalent to anorexia nervosa, obsessive–compulsive dysfunction (OCD) and Tourette syndrome carefully clustering collectively92,93 (Fig. 3e, left dendrogram). For instance, we additional decided the 15 mobile elements that had the strongest brain-specific relationships with OCD, all however one in all which had been associated or particular to neurons (Fig. 3e, proper dendrogram, and Methods). The vast majority of these mobile elements had few genes in widespread with the genes confidently related to OCD (Fig. 3e, orange warmth map; Jaccard indices < 0.04). Nevertheless, after eradicating the few genes confidently related to OCD by means of GWAS, we discovered that the majority elements had been nonetheless enriched with or contained OCD-related genes (Fig. 3e, purple–inexperienced warmth maps), that’s, drug targets for OCD (odds ratio = 8.4 ± 1.8; ChEMBL scientific stage 2 or larger)94, genes associated to OCD from mouse deletion phenotypes (odds ratio = 4.8 ± 0.8; Worldwide Mouse Phenotyping Consortium (IMPC) rating ≥ 0.5)95 or genes much less confidently linked to OCD by means of GWAS (odds ratio = 1.6 ± 0.2, OTAR L2G rating < 0.5) (Methods). Furthermore, these 15 elements with the strongest OCD relationships within the mind had been extra strongly enriched with OCD-related genes than different mobile elements that contained OCD-linked genes (P = 4.3 × 10−11 (drug targets), 3.4 × 10−15 (genes associated to OCD in mice) and 6.6 × 10−7 (genes much less confidently linked to OCD by means of GWAS), in keeping with one-sided Mann–Whitney U-tests).
Collectively, the outcomes above reveal that the proposed relationship scores can prioritize mobile elements which can be enriched with trait-relevant genes. Analogously, we discovered that we may use the connection scores for reconstructing the hierarchical group of the cell, together with maps of subcellular buildings and modules of tissue-specific relations between mobile elements (Supplementary Fig. 19). These observations reveal the potential for our affiliation atlas to facilitate the systematic mapping of relations amongst traits, mobile compartments and sure different ontology phrases.
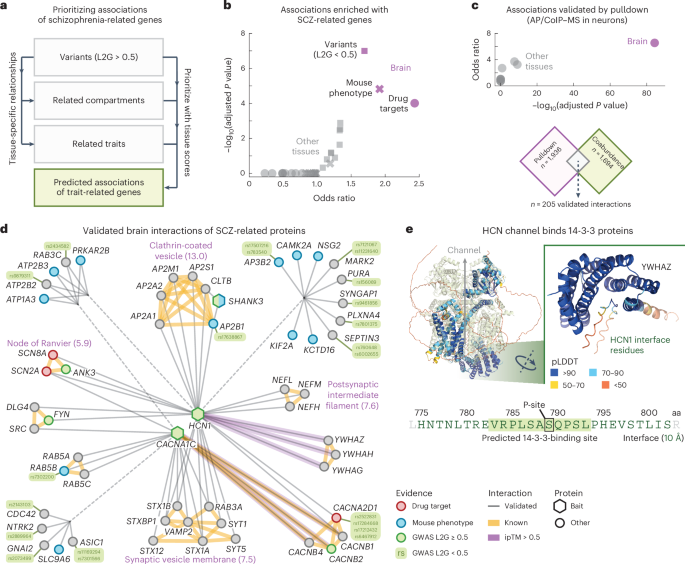
Validated mind interactions for schizophrenia-related genes
The outcomes above point out that the tissue-specific associations may facilitate the prioritization of disease-linked genes by purposeful affiliation. Certainly, direct interactors of disease-linked genes have been used to prioritize causal genes in genetically linked loci and proven to be enriched in profitable drug candidates96,97,98. To discover this in additional element, we constructed a community of mind interactions for schizophrenia (SCZ)-related genes. Particularly, we sought to prioritize extremely ranked associations for the mind that contain SCZ-related genes and which have further proof from orthogonal methodologies.
We began by taking n = 369 genes related to SCZ by means of GWAS research (‘beginning genes’, L2G scores ≥ 0.5) and computed the highest 25 traits and mobile elements that had the strongest tissue-specific relation to SCZ in every tissue (Fig. 4a and Methods). This gave us a set of genes associated to SCZ in a tissue-specific method. For every tissue, we then filtered for protein pairs that had one SCZ beginning gene and one SCZ-related gene and required these protein pairs to have affiliation scores exceeding the 97th percentile of the tissue scores (affiliation rating = 0.70 ± 0.01, 0.73 within the mind), resulting in tissue-specific networks of associations for SCZ-related genes (Supplementary Fig. 20 and Supplementary Desk 8). After eradicating the SCZ beginning genes from the mind community, the remaining genes had been nonetheless enriched for genes related to SCZ in mice (Benjamini–Hochberg (BH)-adjusted P worth = 1.5 × 10−5, IMPC rating ≥ 0.5), drug targets for SCZ (BH-adjusted P worth = 9.8 × 10−5, ChEMBL scientific stage 2 or larger) and different variants related to SCZ (BH-adjusted P worth = 1.0 × 10−7, OTAR L2G scores < 0.5), in keeping with one-sided Fisher actual exams. This enrichment was particular to the mind in comparison with any of the opposite tissues, suggesting that the proposed methodology presents a scientific strategy for prioritizing illness genes of tissue-specific traits (Fig. 4b).
a, Schematic of strategy. Genetic variants related to SCZ are used along with relationships between traits and mobile elements and the tissue scores to prioritize associations for SCZ-related genes. b, Enrichment of predicted associations with genes associated to SCZ by means of mouse phenotypes (IPMC; crosses), drug targets (scientific stage 2 or larger; circles) or GWAS variants (L2G scores < 0.5; squares). c, Enrichment of pulldown interactions within the predicted associations for SCZ-related genes. In b,c, purple symbols characterize the mind and grey symbols characterize different tissues. Scatter plots present the conditional odds ratios and BH-adjusted P values, in keeping with one-sided Fisher actual exams. d, Simplified community of validated mind interactions for SCZ-related genes (Methods). Round and hexagonal nodes had been prey and bait proteins within the pulldown research, respectively. Nodes are coloured as GWAS variants (inexperienced), drug targets (purple), related to SCZ in mice (blue) or different (grey). Grey edges had been predicted from affiliation scores within the mind and validated by means of pulldown experiments. Yellow edges are recognized interactors (bodily associations in STRING; scores > 750). Purple edges have ipTM scores > 0.5. Purple labels annotate subgraphs of recognized interactors with essentially the most enriched GO mobile elements (exponent of BH-adjusted P worth between brackets, in keeping with a one-sided Fisher actual take a look at). e, AlphaFold2 mannequin of the interface between HCN1 and 14-3-3 proteins. Proven are the HCN advanced (Protein Information Financial institution 6UQF; mild inexperienced) aligned with the AlphaFold2 mannequin of HCN and YWHAZ. The sequence exhibits the 14-3-3-binding website of HCN1 in keeping with the AlphaFold2 fashions (inexperienced textual content, 10-Å cutoff; inset), overlaid with the expected 14-3-3-binding website (inexperienced field, 14-3-3-Pred rating = 0.457) and phosphorylation website S789 (black field). Interface residues are coloured by predicted local-distance distinction take a look at.
Certainly, for brain-related problems, together with autism, consideration deficit hyperactivity dysfunction (ADHD), Tourette syndrome and others, we discovered that the proposed methodology of prioritizing protein associations for disease-linked genes enriched, particularly within the mind, for different genes related to the respective dysfunction by means of mouse phenotypes or drug targets, with many additionally being enriched for GWAS variants which can be much less confidently linked to the dysfunction (Supplementary Fig. 21).
Having established that the introduced methodology prioritizes protein associations that enrich for disease-related genes in a tissue-specific method, we sought to validate the prioritized protein associations by means of further proof. Particularly, to additional validate the networks of predicted associations for SCZ-related genes, we assembled a curated dataset of mind interactions established experimentally by means of pulldowns utilizing human mind cells (that’s, AP–MS or coimmunoprecipitation–MS in human microdissected mind tissue or human induced pluripotent stem cell-derived neurons99,100,101,102,103). This dataset contained 7,887 human mind interactions for 30 bait proteins and has been included into the IntAct database9 (Supplementary Desk 9). We filtered these mind interactions for the bait proteins that had been related to SCZ (OTAR L2G rating ≥ 0.5) and additional filtered the tissue-specific networks of SCZ-related genes for associations involving at the least one bait protein. The remaining associations of SCZ-related genes had been strongly enriched with the interactions from pulldowns with SCZ-related baits, particularly for the mind (log BH-adjusted P worth = 84.3, in keeping with a one-sided Fisher actual take a look at) in comparison with the opposite tissues (log BH-adjusted P worth = 1.8 on common) (Fig. 4c). Thus, the mind associations of SCZ-related genes however not the opposite tissues had been experimentally validated by pulldown research and enriched with SCZ-related affiliation companions.
Subsequent, we filtered the prioritized mind associations for SCZ-related genes for interactions that had been additionally discovered by means of the pulldown research to acquire a community of 205 validated mind interactions for SCZ-related genes (common affiliation scores = 0.86 within the mind), which we simplified to solely present synaptic (SynGO) genes having prior proof (Fig. 4d, Methods and Supplementary Desk 10). The visualized community contained 56 proteins linked to a few bait proteins by means of a set of 66 validated mind interactions. These linked proteins included SCZ drug targets (3 proteins; scientific stage 2 or larger), proteins related to SCZ in mice (12 proteins; IMPC rating ≥ 0.5) or proteins linked with SCZ with weaker prior proof (15 proteins; OTAR L2G scores < 0.5). Surprisingly, solely 4 of the visualized mind interactions had been confidently reported in any of the foremost protein interplay databases earlier than our curation effort (CACNA1C with CACNB1, CACNB2 and CACNB4 (STRING scores > 750) and SRC (Signor)) and solely the interactions of SHANK3 with AP2B1 and CLTB had been quantified in additional than three different tissues (affiliation scores = 0.34 ± 0.08 and 0.29 ± 0.04 within the different tissues). These observations help our earlier evaluation that the community of validated interactions of SCZ-related genes is restricted to the mind.
The visualized community contained a number of teams of extremely interconnected proteins (STRING scores > 750). These teams had been enriched with genes of mobile elements typical for neuronal functioning and SCZ, equivalent to a bunch of proteins for the postsynaptic cytoskeleton104 (BH-adjusted P worth = 2.3 × 10−8) or for clathrin-coated vesicles (9.4 × 10−14), in keeping with one-sided Fisher actual exams. For the clathrin vesicle coat, the community linked all subunits of the AP2 advanced and clathrin proteins to HCN1. Apparently, earlier pulldown research confirmed that HCN channels straight work together with TRIP8b (refs. 105,106). TRIP8b regulates the trafficking of HCN channels106 and primarily associates with the AP2 advanced107. Furthermore, whereas AP2 and clathrin will not be cell sort particular, each HCN1 and TRIP8b had been discovered to be enriched at parvalbumin (PV)-positive synapses108, with HCN channels being particular to PV neurons and vital for his or her excessive firing frequencies109,110. Given the hyperlink of PV neurons with SCZ111,112,113,114,115, these observations recommend that AP2 and clathrin could also be concerned in a PV neuron-specific disruption of HCN channel trafficking with SCZ.
To recommend putative interface fashions for the validated mind interactions for SCZ-related genes, we used AlphaFold2 to foretell the buildings for 205 protein interactions, together with your complete visualized community (Fig. 4d and Supplementary Desk 11). The anticipated fashions had extra assured interactions in comparison with fashions for recognized advanced members from CORUM116 (common pDockQ scores = 0.20 and 0.13, respectively, P = 1.6 × 10−19, in keeping with a one-sided Mann–Whitney U-test). In whole, we recognized 15 moderate-confidence interactions (interface predicted template modeling (ipTM) scores > 0.5). These included the brain-specific binding of all three 14-3-3 proteins (YWHAG, YWHAH and YWHAZ) with HCN1 (Fig. 4e; common affiliation rating = 0.82, common ipTM rating = 0.65). The interfaces of those three fashions overlap and are situated within the C-terminal disordered area of HCN1 (residues 775–802). This consensus interface features a predicted 14-4-3-binding website (centered round S789; common ipTM rating = 0.75 on the putative binding website, 14-3-3-Pred rating = 0.457)117 that has been verified by means of pulldown experiments118. Certainly, the binding of 14-3-3 proteins with HCN1 was discovered to be depending on the phosphorylation of S789, with the interplay between 14-3-3 and HCN1 probably inhibiting HCN1 degradation118.
Lastly, the community contained 15 genes inside loci genetically related to SCZ that had weaker proof supporting them because the causal genes at every locus. Given their interplay with different SCZ-related genes, these could possibly be prioritized as extra probably causal due to their purposeful roles. A few of these genes (AP2B1, ATP2B2 and SYNGAP1) had the very best L2G rating for his or her respective locus with single-nucleotide polymorphisms (SNPs) linked to SCZ however had scores under the 0.5 cutoff used (0.457, 0.264 and 0.251, respectively)90,91. Along with the AP2 advanced, we discovered a member of the AP3 advanced (AP3B2). AP3B2 had the second highest rating for the locus with SNPs (rs783540), which had splicing and expression quantitative trait locus (QTL) associations with AP3B2 however was ranked larger for disruption of CPEB1 on condition that the variant lies inside a CPEB1 intron. Equally, MARK2 was ranked second for 2 SNPs (rs7121067 and rs11231640), each having splicing and promoter seize Hello-C associations with MARK2 however being ranked larger for disrupting RCOR2 due to proximity to its transcription begin website. CDC42 and NTRK2 had the second highest L2G scores for his or her locus however the high related genes (WNT4 and AGTPBP1, respectively) had decrease and fewer particular expression within the mind12.
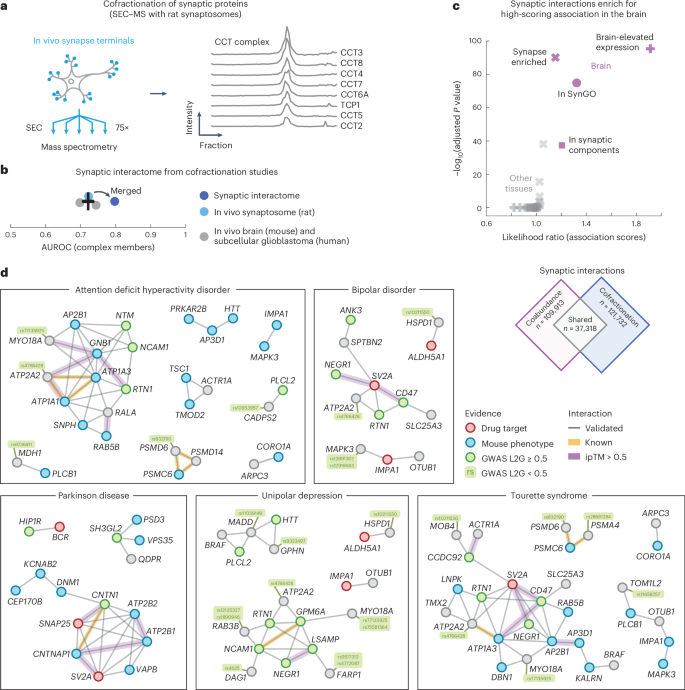
Cofractionation-derived synapse-specific interactome
As a closing software of our protein affiliation atlas, we centered on the interactome of synapses. To take action, we ready and purified synaptosomes from rat brains as an orthogonal strategy for validating the mind associations. We fractionated the synaptosomes with size-exclusion chromatography (SEC) into 75 fractions that had been then subjected to liquid chromatography (LC)–MS/MS. A complete of three,409 distinctive proteins had been detected, together with well-known protein complexes such because the CCT advanced subunits, whose profiles correlate throughout the fractions (Fig. 5a; common correlation coefficient = 0.96).
a, Schematic of experiment. In vivo synaptosomes (mild blue) from rat neurons had been purified and fractionated into 75 fractions by means of SEC and subjected to LC–MS/MS. Elution profiles present the protein intensities from MS for the CCT advanced members (proper). b, AUC values for the cofractionation research (in vivo mouse mind, grey; subcellular glioblastoma, grey; in vivo rat synaptosome, mild blue) and for the merged synaptic interactome (darkish blue). Positives had been outlined by advanced members in CORUM. The error bar exhibits the imply and s.e.m. (n = 3). c, Comparability of affiliation scores of interactions between synaptic proteins or interactions of different proteins within the mind (purple) and the opposite tissues (grey). Interactions are from the synaptic interactome (rating > 0.8). Information had been derived utilizing a one-sided Mann–Whitney U-test, BH-adjusted P values and median probability ratios. Synaptic proteins included people who had been enriched within the synapse (crosses), reported in SynGO (circles), related to GO synaptic elements (squares) or had brain-elevated expression in keeping with The Protein Atlas (pluses). d, Networks of validated interactions between synaptic proteins (SynGO or enriched in mouse tissues) associated to ADHD, bipolar dysfunction, Parkinson illness, unipolar despair or Tourette syndrome. Nodes are coloured by affiliation to the trait by means of GWAS (inexperienced), drug targets (purple), mouse phenotypes (blue) or different (grey) (Methods). Grey edges had been predicted from affiliation scores within the mind and validated by means of the cofractionation research. Yellow edges are recognized interactors. Purple edges have ipTM scores > 0.5.
We preprocessed the fractionation profiles of the synaptosome and computed the coabundance of proteins throughout fractions to attain the cofractionation of 4,276,350 protein pairs in rat synapses (Methods). To extend the boldness of the interplay scores, we then mixed the rat synaptosome with different cofractionation profiles from in vivo mouse brains6 and subcellular fractionation profiles from human glioblastoma cells7. We merged the cofractionation research by orthologs that had been quantified in all three datasets and computed interplay chances with a logistic mannequin utilizing the CORUM database as positives (Methods and Supplementary Desk 12). The ensuing synaptic interactome quantified 1,309,771 interplay chances for 1,619 proteins and improved the restoration of recognized interactions in comparison with the cofractionation research individually (Fig. 5b; AUC = 0.80 and 0.72). Of the 1,619 proteins within the interactome, 24% are annotated as synaptic proteins within the SynGO database, 49% have been reported as synapse-enriched in mouse brains and 56% have beforehand been recognized by means of crosslinking MS (XL-MS) of the mouse synaptosome84,108,119. All XL-MS interactions for these proteins had been quantified in our synaptic interactome, having affiliation scores of 0.59 in comparison with 0.37 for different associations (P = 2.0 × 10−44, in keeping with a one-sided Mann–Whitney U-test). Furthermore, the synaptic interactions between synapse-enriched proteins had been extra probably in comparison with different interactions (common affiliation scores = 0.40 and 0.36, P < 1 × 10−300, in keeping with a one-sided Mann–Whitney U-test). Collectively, these observations recommend that our synaptic interactome largely aligns with present state-of-the-art synaptic interplay assets and scores the probability of interactions for a variety of synaptic proteins, with interactions being extra probably for synaptic proteins in comparison with different proteins.
Validated synaptic interactions for mind illness genes
The interactions between synapse-enriched proteins fashioned extra probably associations in comparison with associations of nonsynaptic proteins, particularly for the mind in comparison with the opposite tissues of our affiliation atlas (Fig. 5c). Given this brain-specific elevated probability of synaptic interactions, we constructed a community of interactions between synaptic proteins (in SynGO or synapse-enriched in mouse brains) that had been each probably coabundant within the mind (109,913 associations) and sure cofractionated for the synaptosome (121,732 interactions). The ensuing community consisted of a set of 37,318 validated protein interactions between synaptic proteins (Supplementary Desk 13). These synaptic interactions had been primarily particular to the mind as a result of few of the interactions had been probably for almost all of different tissues in our affiliation atlas (20%) and solely a small fraction have been reported in any protein interplay database (5.8% in STRING, 1.3% in HuMAP, 3.6% in IntAct and 1.6% in BioPlex).
We had been significantly within the validated interactions of synaptic proteins which can be related to mind problems. As earlier than, we filtered for GWAS traits that had related genes by means of mouse phenotyping (IMPC) or recognized drug targets (ChEMBL) and whose trait-level affiliation scores had been elevated within the mind in comparison with the opposite tissues (z rating > 1; Methods). We discovered that 10 of the ensuing 13 traits had been problems clearly associated to the mind and chosen the 727 assured synaptic interactions between genes related to these brain-specific traits (affiliation scores = 0.7 and 0.81 on common within the synaptosome and mind, respectively). Moreover, to recommend putative interface fashions, we used AlphaFold2 to foretell the buildings for these 727 interactions. The anticipated fashions had extra assured interactions in comparison with fashions for recognized interactors in CORUM (common pDockQ scores = 0.28 and 0.13, P = 3.7 × 10−147, in keeping with a one-sided Mann–Whitney U-test) or HuMAP (pDockQ scores = 0.28 and 0.25, P = 2.4 × 10−17) and high-confidence fashions (ipTM scores > 0.7) had been enriched for added proof from XL-MS experiments in mouse synaptosomes119 (Supplementary Fig. 22). In whole, we recognized 105 moderate-confidence interactions (ipTM scores > 0.5; Supplementary Desk 14). Lastly, we visualized (simplified) the networks of validated synaptic interactions between trait-related genes of the mind problems (Fig. 5d and Methods).
Prioritizing synaptic illness genes for mind problems
A number of genes within the networks had weaker prior proof supporting them because the causal genes at loci genetically related to the mind problems. These genes could possibly be prioritized as extra prone to be causal for the problems due to their validated synaptic interactions with different genes that had been confidently related to the identical problems. As earlier than, we seemed for genes with the very best (under cutoff) L2G scores for his or her respective loci with SNPs linked to the mind problems. We discovered probably causal genes for ADHD (MDH1, rating = 0.491; CADPS2, rating = 0.340; PIK3C3, rating = 0.323), SCZ (TOM1L2, rating = 0.492; AP2B1, rating = 0.457; PSD3, rating = 0.429; MYO18A, rating = 0.428; ATP2B2, rating = 0.264; TMX2, rating = 0.232), Alzheimer illness (CLPTM1, rating = 0.378; MADD, rating = 0.315), autism (ATP2B2, rating = 0.264; ATP2A2, rating = 0.243), unipolar despair (MADD, rating = 0.244) and bipolar dysfunction (ATP2A2, rating = 0.206), with the final variant (MYO18A, rating = 0.428) moreover being probably causal for Tourette syndrome, OCD, ADHD and unipolar despair90,91. Of those genes, all however ATP2A2, MDH1, MYO18A and PIK3C3 had the very best or second highest expression in mind tissue12.
Moreover, we discovered genes with synaptic interactions that didn’t have the very best L2G rating for the respective SNP linked to the mind problems however nonetheless had further proof. For instance, we discovered that PAFAH1B1 related to RHOA in our synaptic interactome, with each having weak prior proof for being causal to despair. PAFAH1B1 has a task in neural mobility and is required for activation of Rho guanosine triphosphatases equivalent to RhoA120. PAFAH1B1 was the second highest scoring gene for the locus with an SNP (rs12938775) linked to unipolar despair. This variant has expression QTL associations with PAFAH1B1 and lies inside a PAFAH1B1 intron, regardless of being extra distant to the transcription begin website of PAFAH1B1 in comparison with the very best scoring gene (CLUH). Nevertheless, PAFAH1B1 encodes a synaptic protein, whereas CLUH doesn’t, with the expression of PAFAH1B1 being larger and extra particular to the mind in comparison with CLUH12. General, this instance demonstrates how orthogonal approaches focusing on subcellular buildings and particular person tissues can present tissue-specific protein interplay networks and support within the prioritization of genes prone to be causal for tissue-related problems.