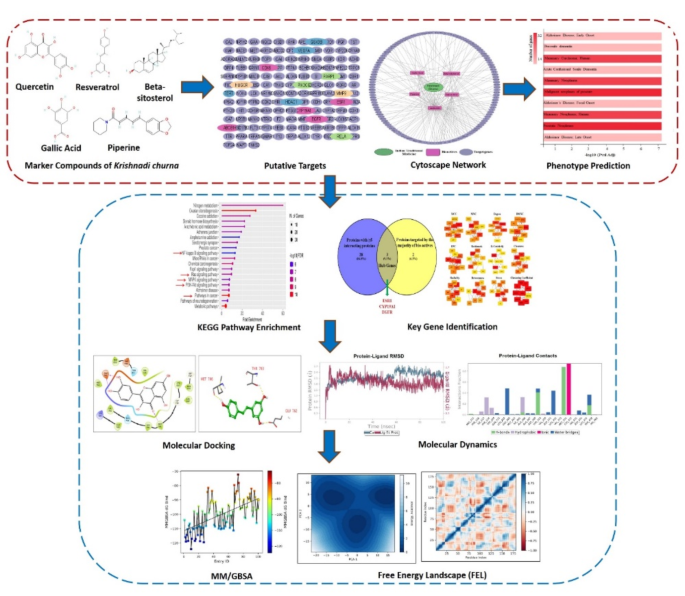
5 marker compounds-gallic acid (phenol), piperine (alkaloid), resveratrol (phenol), quercetin (flavonol), and beta-sitosterol (phytosterol), have been checked and quantified earlier for his or her presence in Krishnadi Churna by HPTLC technique17. This research goals to search out out the putative human targets and related phenotype of those 5 marker compounds. All 5 bio-actives have been screened for Oral bioavailability, Drug-likeness, and pharmacokinetic evaluation utilizing SwissADME (Desk 1 and Supplementary Fig. 1); besides Beta-sitosterol, all certified for Drug likeness primarily based on predefined standards, and amongst all solely Piperine certified for oral bioavailability. An in depth description of ADME screening and pharmacokinetic evaluation is proven in Supplementary File 2. Since all 5 bio-actives are the marker compounds of polyherbal formulation, these predictions could change with the presence of a number of phytoconstituents collectively. Therefore, all 5 biomarkers have been thought of for additional evaluation.
Targets and community evaluation of polyherbal marker compounds
Putative targets for every bioactive have been predicted utilizing BindingDB with a similarity rating of 0.7.; the place 29 targets for Resveratrol, 14 for Gallic Acid, 8 for Piperine, 90 for Quercetin, and 27 for Beta-sitosterol have been discovered. Within the group of any two bio-actives; Resveratrol and Galic Acid had 5, Resveratrol and Quercetin had 15, Resveratrol and Beta-sitosterol had 2, Gallic Acid and Quercetin had 6, Piperine and Quercetin had 3, and Quercetin and Beta-sitosterol had 7 widespread targets. Among the many group of any three bio-actives, solely Resveratrol, Gallic Acid and Quercetin had 3, and Resveratrol, Quercetin and Beta-sitosterol had 2 widespread targets. No widespread goal was discovered for any 4 or 5 bioactive teams. All of the bioactives together with their goal checklist are proven in Desk 2.
A complete of 168 compound targets have been discovered and after eradicating duplicates the remaining 135 targets have been eloquently represented together with their bio-actives utilizing Cytoscape (Fig. 2). A Cytoscape community was additionally constructed for particular person bioactive and their targets (Supplementary Fig. 2). Additional, a Protein-protein interplay (PPI) community was constructed utilizing STRING, and the targets with an interplay rating of 0.9, the very best confidence is proven in Fig. 3. Close to about 67.4% of the whole putative targets had the very best confidence of interplay, just a few of those proteins are HDAC1, ESR1, VEGFA, RELA, EGFR, and many others.
Polyherbal marker compounds goal cancer-related pathways
To elucidate the phenotype of goal genes, a listing of goal genes was uploaded as an enter in GeneCodis4, annotation for the phenotype (DisGeNET, HPO, OMIM) was chosen, and evaluation was launched. As proven in Fig. 4a., most genes confirmed the phenotype for mammary carcinoma, adopted by prostate most cancers and Alzheimer’s Illness. Determine 4b. represents the annotations and the genes related to them within the type of a community the place genes like RELA, ABCB1, CYP19A1, SERPINE1, AKR1C3, MMP9, TERT, and many others., belonged to mammary and prostate most cancers, and genes like APP, BACE1, MOAB, and many others. belonged to Alzheimer’s illness; whereas ESR1, EGFR, VEGFA, GSK3B, ACHE, and many others. are essential for most cancers and Alzheimer’s each. Additional, KEGG pathway enrichment evaluation25,26,27,28 was carried out and as proven in Fig. 4c, Pathways in most cancers, NF-kappa B signaling pathway, Ras signaling pathway, MAPK signaling pathway, and PI3K-Akt signaling pathway have been among the many extremely enriched pathway. Curiously, these are the important thing pathways for breast most cancers survival, proliferation, metastasis, and drug resistance. A myriad of experiences is obtainable showcasing their essential position in breast most cancers development and the way totally different methods are being tailored to focus on these pathways37,38,39,40,41,42. Gene Ontology (GO) evaluation was additionally carried out on course genes and as proven in Fig. 4d and Supplementary Fig. 3, these potential targets of polyherbal marker compounds have been primarily positioned within the cytoplasm (GO:0005737), receptor complicated (GO:0043235), cell floor (GO:0009986), nucleoplasm (GO:0005654), protein kinase complicated (GO: 1902911), chromosome (GO: 0005694); concerned within the organic course of like cell dying (GO:0008219), apoptotic course of (GO:0006915), peptidyl-serine modification (GO:0018209), cell inhabitants proliferation (GO:0008283), endocytosis (GO:0006897), cell cycle (GO:0007049), cell junction meeting (GO:0034329); and regulates molecular capabilities like similar protein binding (GO:0042802), nuclear receptor exercise (GO:0004879), signaling receptor exercise (GO:0038023), enzyme binding (GO:0019899), protein-containing complicated binding (GO:0044877), chromatin binding (GO:0003682), non-membrane spanning protein tyrosine phosphatase exercise (GO:0004726), and many others.
(a) Bar graph displaying essentially the most important phenotype for natural phytoconstituents’ targets. (b) Community of annotations and genes related to the gene targets’ phenotype. (c) KEGG Pathway enrichment evaluation of BC genes focused by natural phytoconstituents. This graph was generated utilizing ShinyGO 0.7624,25,26,27,28. (d) Gene Ontology evaluation displaying Molecular Perform, Organic Course of and Mobile Part of gene targets.
Key genes recognized are essential for BC development
Additional, key genes have been recognized among the many numerous targets of marker compounds. To begin with, 28 targets with ≥ 5 interacting proteins within the PPI community have been chosen. Concurrently, proteins focused by the vast majority of bio-actives have been chosen, right here, as evident from Desk 2, 5 proteins-ESR1, CA12, CYP19A1, EGFR, and TTR have been focused by 3 bioactive compounds and therefore chosen for evaluation. Additional, the widespread targets from the 2 picks have been thought of as the important thing genes (Fig. 5a). Three key genes chosen have been, ESR1, CYP19A1, and EGFR; loads of experiences primarily based on scientific and pre-clinical research have proven the essential position of those genes in poor scientific outcomes, relapse of illness, remedy resistance and metastasis of BC43,44,45,46. Additionally, primarily based on the calculation of cytoHubba, the highest 10 nodes ranked by all twelve algorithms have been evaluated. Out of 12 algorithms, ESR1 was ranked as the highest 10 nodes in 9 algorithms and EGFR in 10 algorithms (Fig. 5b) therefore thought of because the tremendous essential gene on this research.
Additional, the Human Protein Atlas was explored to verify the 5-year survival of Breast most cancers sufferers with diversified expression of the important thing genes- ESR1, CYP19A1, and EGFR18. The five-year survival fee for sufferers with excessive CYP19A1 expression was 76%, in comparison with 83% for these with low expression, with a p-value of 0.094. Equally, for ESR1, the survival charges have been 80% and 82% respectively, with a p-value of 0.083. For EGFR, the charges have been 85% and 80% with a p-value of 0.026. Curiously, a constructive correlation of their expression was noticed with poor survival of BC sufferers (Fig. 5c–e). Nonetheless, EGFR confirmed an reverse development, which might be due to its conflicting position in BC47. Moreover, we checked the mutation frequency of the goal gene in BC sufferers utilizing this system and instruments made out there on-line at cBioportal (cBioPortal for Most cancers Genomics)19,20. Curiously, out of 135 putative targets, 134 genes have been discovered mutated in breast most cancers sufferers (Fig. 5f). Additional, the mutation frequency of key genes was recognized in breast most cancers sufferers, and as proven in Desk 3, 7.3% mutation frequency was noticed for ESR1, 1.5% for EGFR and 0.30% for CYP19A1. General, the printed manuscripts43,44,45,46 and database outcomes spotlight the essential position of those genes in breast most cancers development and prognosis, underscoring the significance of growing therapeutic methods that concentrate on them.
(a) Venn Diagram depicting the important thing genes recognized among the many putative targets of natural phytoconstituents. (b) Recognized Hub gene community of natural phytoconstituents for various algorithms of CytoHubba. Survival Plot with excessive and low expression of (c) ESR1, (d) CYP19A1, (e) EGFR in BC sufferers from Human Protein Atlas on-line database. (f) Venn Diagram depicting mutation standing of goal genes in BC from cBioportal.
Molecular docking
Marker compounds have been predicted to own a great affinity for key genes (targets). Potential areas within the goal receptor ESR1, CYP19A1, and EGFR have been examined to search out the potential binding poses of bioactive, utilizing subunits of the protein construction ESR1(5T97), CYP19A1 (4GL7), and EGFR (4LRM). Gallic acid (ID 370), Piperine (ID 638024), Quercetin (ID 5280343), and Resveratrol (ID 445154) have been docked with the above targets. Raloxifene Core (ID 445920), Letrozole (ID 3902), and Erlotinib have been the constructive controls for ESR1, CYP19A1, and EGFR respectively. Outcomes obtained after docking revealed sturdy interactions of bioactive in opposition to the targets when it comes to binding power (Kcal/mol) proven in Desk 4, least binding power (extra detrimental binding power) is interpreted as extra stability.
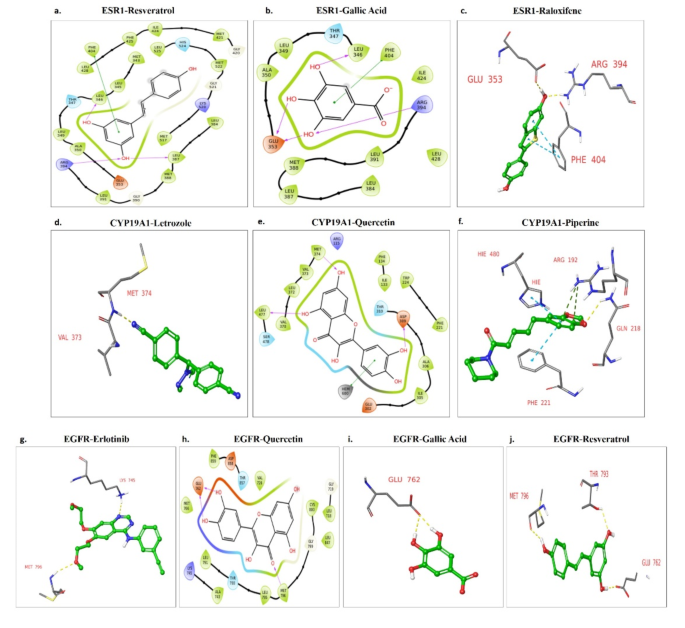
The binding affinity of ESR1 (5T97) for constructive management (Raloxifene Core) was − 8.819 Kcal/mol. Piperine expressed − 8.288 Kcal/mol binding affinity with 5T97. It was nearest to the constructive management. Resveratrol (-7.95 Kcal/mol), Gallic acid (-6.984 Kcal/mol), and Quercetin (-6.755 Kcal/mol) additionally exhibited a good binding affinity. Resveratrol confirmed hydrogen bonding with LEU387, ARG394, LEU346 and pi-pi bond with PHE404 (Fig. 6a). Gallic acid confirmed Hydrogen bonding with LEU346, GLU353, ARG394, and pi-pi bonding with PHE404 (Fig. 6b). With twin hydrogen bonding with ASP351, the Quercetin molecule has no different bond in molecular docking. Raloxifene core reveals hydrogen bonding with GLU353, and ARG394 whereas pi-pi bonding with PHE404 (Fig. 6c).
The binding affinity of CYP19A1 (4GL7) for constructive management (Letrozole) was − 6.771 kcal/mol. Letrozole reveals hydrogen bonding with VAL373 and MET374 (Fig. 6d). Amongst bioactive, Quercetin reveals one of the best docking rating (-8.628 Kcal/mol) and hydrogen bonds ASP309, MET374, and LEU477 and a pi-pi bonding HEM600(Fig. 6e). Resveratrol (-7.089 Kcal/mol), Piperine (-5.878 Kcal/mol), and gallic acid (-5.516 Kcal/mol) additionally mirrored a average binding affinity. Piperine has a hydrogen bond with GLN218, pi-cation bond with ARG192 and pi-pi Bonding with HIE480 and PHE221 (Fig. 6f).
The binding affinity of EGFR (4LRM) with constructive management (Erlotinib) was − 6.285 Kcal/mol. Compared, the binding affinity for bioactive was Quercetin (-8.629 Kcal/mol), Resveratrol (-7.772 Kcal/mol), Gallic acid (-6.501 Kcal/mol), and Piperine (-6.464 Kcal/mol). All ligands exhibit higher docking scores than the constructive management. Erlotinib has hydrogen bond interplay at MET796 and LYS745 (Fig. 6g). Quercetin has hydrogen bonds with MET796 and GLU762 (Fig. 6h). Gallic acid confirmed 2 converging hydrogen bonds with GLU762 (Fig. 6i). With the very best interplay, Resveratrol reveals 3 hydrogen bonding with MET796, THR793, and GLU762 (Fig. 6j).
The mutant variants of ESR1 and EGFR have been additionally analyzed to determine the potential binding poses of the bioactive compounds utilizing the protein subunits 4Q50 ESR1 (D538G) and 4I20 EGFR (L858R). Docking outcomes revealed sturdy interactions between the bioactives and the goal proteins, as indicated by their binding energies (Kcal/mol) introduced in Desk 5.
The binding affinity of Raloxifene core, the constructive management, with the mutant ESR1 (D538G) was recorded at -10.116 Kcal/mol. Compared, Gallic acid (-6.331 Kcal/mol), Piperine (-8.769 Kcal/mol), Quercetin (-6.894 Kcal/mol), and Resveratrol (-8.149 Kcal/mol) additionally confirmed important binding affinities. When it comes to molecular interactions, Raloxifene core confirmed a hydrogen bond with GLU353 and a pi-pi bond with PHE404 (Fig. 7a). Gallic acid fashioned hydrogen bonds with GLU353, LEU387, and a pi-pi bond with PHE404 (Fig. 7b). Piperine and Quercetin exhibited hydrogen bonding with HIS524 and ACE532, respectively, whereas Resveratrol fashioned hydrogen bonds with LEU387 and GLU353, together with a pi-pi bond with PHE404 (Fig. 7c-e).
The binding affinity of mutant EGFR (L858R) with the constructive management, Erlotinib, was discovered to be -4.456 Kcal/mol. Notably, all of the examined bioactives demonstrated superior docking scores in comparison with the constructive management: Gallic acid (-5.177 Kcal/mol), Piperine (-4.673 Kcal/mol), Quercetin (-5.686 Kcal/mol), and Resveratrol (-5.915 Kcal/mol). When it comes to particular interactions, Erlotinib confirmed hydrogen bonding with CYS797 and LYS745. Gallic acid fashioned hydrogen bonds with ASP800 and CYS797, whereas Resveratrol confirmed hydrogen bonding with LYS745, MET793, and ASP800. Quercetin established hydrogen bonds with ASN842 and ASP800. Piperine exhibited a hydrogen bond with CYS797(Fig. 7f-j).
Docking outcomes displaying H-bonds and different interactions between protein-ligand complicated of (a) ESR1-resveratrol, (b) ESR1-gallic acid, (c) ESR1-raloxifene core, (d) CYP19A1-letrozole, (e) CYP19A1-quercetin, (f) CYP19A1-piperine, (g) EGFR-Erlotinib, (h) EGFR-quercetin, (i) EGFR-gallic acid, (j) EGFR-resveratrol.
Docking outcomes displaying H-bonds and different interactions between protein-ligand complicated of (a) ESR1 (D538G)-raloxifene core, (b) ESR1 (D538G)-gallic acid, (c) ESR1 (D538G)-piperine, (d) ESR1 (D538G)-quercetin, (e) ESR1 (D538G)-resveratrol, (f) EGFR (L858R)-erlotinib, (g) EGFR(L858R)-gallic acid, (h) EGFR(L858R)-piperine, (i) EGFR(L858R)-resveratrol, (j) EGFR (L858R)-quercetin.
Molecular dynamics
Molecular dynamics simulations have been carried out to research the protein-ligand complicated conduct in specific organic options. Such conduct is predicted utilizing RMSD (root imply sq. deviation), ligand affinity, and non-bonding interplay within the protein-ligand complicated.
One of the best dock rating for CYP19A1 was Quercetin, therefore CYP19A1-Quercetin complicated was chosen for Molecular docking simulation. Advanced simulation was carried out at an ordinary temperature (300 Ok) for a stipulated time of 100 nanoseconds. Protein was positively charged with whole residue of 454. Preliminary protein RMSD fluctuation was almost at 1 A. Ligand was secure between 2 and a pair of.7 A all through the simulation interval. Each the ligand and the protein have been secure in a body of 1–2.7 A. Advanced interplay was secure all through besides at across the fiftieth ns body. Right here it deviates for greater than 0.8 A. Determine 8a reveals the numerous interplay between the ligand and protein. A complete of 25 contacts have been famous as proven in Fig. 8b. The complicated confirmed Hydrogen, Hydrophobic, and ionic bonds, having the very best interplay fraction of 1. Hydrogen bond was famous at ARG_115, ALA_306, THR_310 (39%), MET_374 (87%), SER_478. CYS 437 had interplay with Ferrous ions for 94% of the length of the simulation. Protein SSE was 54.54% with 47.86% of alpha helix and 6.68% of beta strands (Fig. 8c).
Advanced EGFR-Quercetin expressed one of the best dock rating and was additional subjected to simulation at customary temperature (300 Ok) for 100 nanoseconds. The interplay had positively charged protein of 309 whole residues. Preliminary protein RMSD fluctuation was almost at 1 A. Protein was secure all through the simulation time at 2–2.5 A. Ligand was secure besides at round 20–40 nanoseconds. Advanced interplay was not discovered throughout this timeframe as proven in Fig. 8d. The complicated had almost 24 contacts as depicted in Fig. 8e. Advanced reveals H-bond and Hydrophobic bonds, having the very best interplay fraction round 1.6. H bond was seen at LYS_745, GLU_762 (75%), THR_793, GLN_794 (91%), MET_796 (98%), and ASP_858 (52%). Whole protein secondary constructions have been 43.93%, with 28.54% alpha helix and 15.38% beta strands (Fig. 8f).
The ESR1-Piperine protein-ligand complicated expressed one of the best dock rating and was additional subjected to simulation for 100 nanoseconds. Protein was negatively charged with 185 whole residues. Preliminary protein RMSD fluctuation was almost at 1 A. Each protein and ligand have been unstable throughout this framework. Ligand RMSD fluctuated between 6 and 14 A. The complicated had interplay twice in the course of the 100 nanoseconds body, at round 20ns and 60-80ns (Fig. 8g). A complete of 32 contacts was seen between the protein-ligand as proven in Fig. 8h Advanced reveals H-bond and Hydrophobic bonds, having the very best interplay fraction round 0.25. No ionic bonds have been famous. Hydrogen bond was famous at GLY_344, THR_347, TRP_383, GLY_420, MET_421, NMA_525. Not one of the interactions occurred for greater than 30.0% of the simulation time within the chosen trajectory. Whole protein secondary constructions have been 71.32%, with 69.82% alpha helix and 1.50% beta strands (Fig. 8i).
The best docking rating for mutant EGFR-L858R was noticed with Resveratrol, making the EGFR (L858R)-Resveratrol complicated the prime candidate for molecular docking simulation. The simulation was performed at an ordinary temperature of 300 Ok over 100 nanoseconds. The protein, consisting of 289 residues, carried a constructive cost. The preliminary RMSD fluctuation for the protein was round 1Å, whereas the ligand remained secure inside a variety of 1–2Å all through the simulation. Each the ligand and protein exhibited stability between 1-2.4Å. Notably, the complicated interplay remained secure on the tenth and seventieth nanosecond frames. As proven in Fig. 9a, important interactions occurred between the ligand and protein, with a complete of 19 contacts recognized (Fig. 9b). The complicated featured hydrogen, hydrophobic, and ionic bonds, with the very best interplay fraction being 1.4. Hydrogen bonds have been noticed at LYS_745 (55%), GLN_791 (76%), MET_793 (95%), and ASP_800 (40%). The protein secondary construction components (SSE) consisted of 48.20%, with 30.68% alpha helices and 17.52% beta strands (Fig. 9c).
Equally, one of the best docking rating for mutant ESR1-D538G was achieved with Piperine, prompting molecular dynamics evaluation of the ESR1-Piperine complicated. The simulation, carried out at 300 Ok for 100 nanoseconds. The protein, composed of 222 residues, was positively charged. The preliminary RMSD fluctuation for the protein was about 1Å, whereas the ligand remained secure between 4.8–5.6Å all through the simulation. The complicated interplay remained secure at numerous intervals, notably on the tenth, thirtieth, and seventieth nanosecond frames. As illustrated in Fig. 9d, important interactions have been noticed between the ligand and protein, with 25 contacts famous (Fig. 9e). Hydrogen, hydrophobic, and ionic bonds have been current, with the very best interplay fraction recorded at 0.7. Hydrogen bonds have been noticed at GLY_420, HIS_524 (74%), and SER_527. The protein SSE was 67.65%, comprising 64.61% alpha helices and three.04% beta strands (Fig. 9f).
Molecular docking simulation outcomes of (a) protein ligand RMSD, (b) protein ligand contacts and (c) protein secondary construction evaluation of CYP19A1-Quercetin complicated. RMSD, Protein Ligand Contacts and Protein secondary construction evaluation of (d–f) EGFR-Quercetin and (g–i) ESR1-Piperine complicated.
Molecular docking simulation outcomes of (a) protein ligand RMSD (b) protein ligand contacts and (c) protein secondary construction evaluation of EGFR (L858R)-Resveratrol complicated. RMSD, Protein Ligand Contacts and Protein secondary construction evaluation of (d-f) ESR1(D538G)-Piperine complicated.
MM/GBSA evaluation highlights a great ligand-binding affinity
Prime MM/GBSA (Molecular Mechanics Generalized Born Floor Space), additionally known as Gibbs binding free power, is a computational chemistry technique extensively used to estimate the binding free power of small molecules with their goal proteins. This method combines molecular mechanics calculations with the Generalized Born mannequin for solvation, incorporating a floor space time period to account for nonpolar solvation results inside the protein binding web site. By evaluating Gibbs binding free power, Prime MM/GBSA supplies essential insights into ligand-receptor interplay energy, serving to determine promising candidates for drug design.
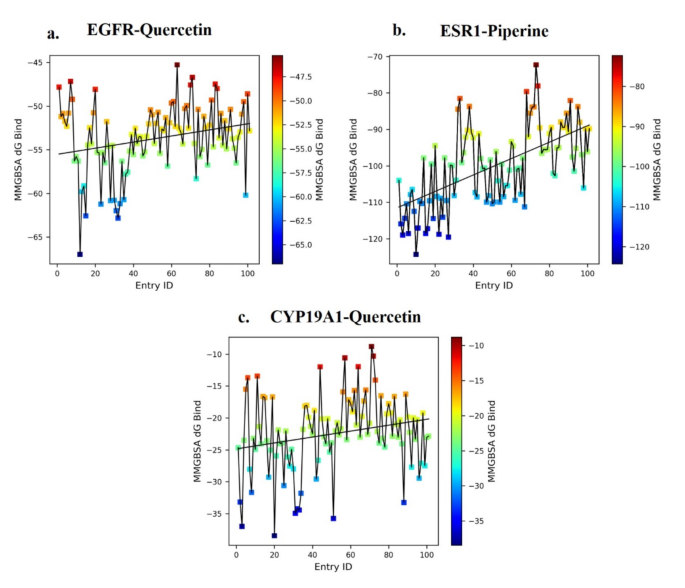
On this research, the binding power evaluation of molecular dynamics (MD) trajectories, taken at fastened intervals of 10 frames throughout 1000 frames for 3 complexes—EGFR-Quercetin (5280343), ESR1-Piperine (638024), and CYP19A1-Quercetin (5280343)—yielded the numerous observations. The EGFR-Quercetin (5280343) complicated exhibited the binding affinity fluctuations inside a variety of -67 kcal/mol at 14 ns to -45 kcal/mol at 65 ns, with intermediate values oscillating between − 56 kcal/mol and − 52 kcal/mol all through the 0-100 ns simulation body (Fig. 10a). This means comparatively secure binding interactions with average power variations over time. Within the ESR1-Piperine (638024) complicated (Fig. 10b), the bottom binding power was − 124 kcal/mol at 10 ns, and the very best was − 72 kcal/mol at 72 ns. The typical binding power, represented by the best-fit line, fluctuated between − 111 kcal/mol and − 90 kcal/mol over the 0-100 ns body, reflecting sturdy and constant binding affinity, with important power fluctuations highlighting dynamic ligand-receptor interactions. For the CYP19A1-Quercetin (5280343) complicated (Fig. 10c), the bottom binding power noticed was − 37 kcal/mol at 20 ns, whereas the very best was − 9 kcal/mol at 71 ns. The power diversified between − 25 kcal/mol and − 20 kcal/mol all through the 0-100 ns simulation, indicating a comparatively weaker interplay in comparison with the opposite complexes.
General, the research reveals that every one three ligands bind to their respective targets, with the ESR1-Piperine (638024) complicated demonstrating the strongest and most secure binding affinity. The EGFR-Quercetin (5280343) complicated additionally reveals average stability, suggesting potential, although additional refinement could also be required for optimization.
Publish molecular dynamics structural evaluation of protein-ligand complicated
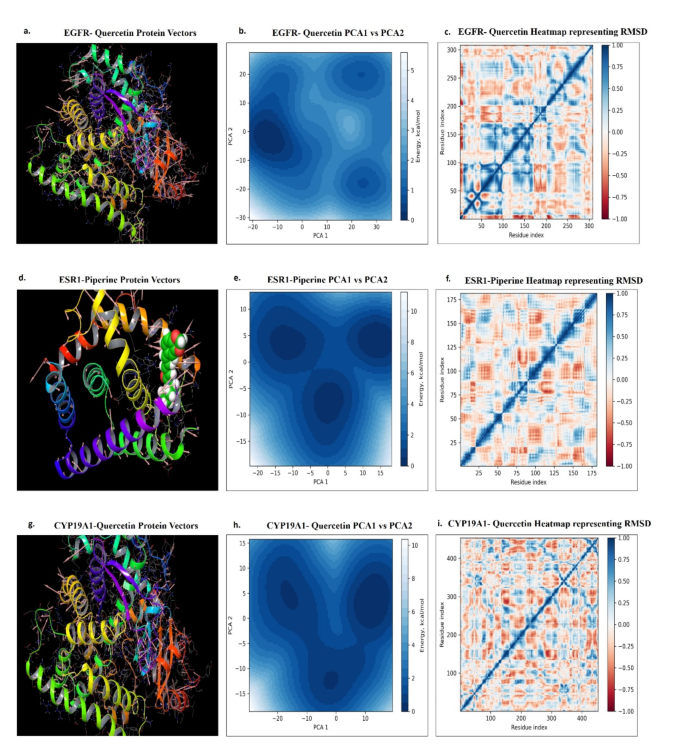
An in-depth molecular dynamics trajectory evaluation utilizing principal element evaluation (PCA) of the EGFR in complicated with the ligand Quercetin (528034) has revealed key computational insights into this interplay. The research targeted on the primary two principal modes, which drive the collective movement of the protein-ligand complicated in the course of the molecular simulations. Pronounced anticorrelated fluctuations, notably within the loop areas of the binding web site residues, have been noticed, depicted by pink vectors within the picture proven as Fig. 11a. These fluctuations recommend that Quercetin’s binding induces coordinated actions inside the EGFR construction, which might be essential for the receptor’s useful dynamics. The EGFR-Quercetin interplay exhibited outstanding stability, with restrained movement of EGFR within the presence of Quercetin, signaling a sturdy binding interplay. This stability was additional supported by the free power panorama (FEL) evaluation, the place the kernel distribution perform served because the third dimension. The FEL plot revealed three distinct power minima, representing secure conformational states of the EGFR-Quercetin complicated (Fig. 11b). Importantly, the structural frames corresponding to those minima didn’t exhibit important conformational modifications, implying that the ligand binding interactions remained largely intact and weren’t disrupted by any main protein structural reconfigurations. Additional evaluation confirmed that the foundation imply sq. deviation (RMSD) of Quercetin within the EGFR-Quercetin (528034) complicated ranged round 1.2 Å, with the radius of gyration (rGyr) spanning from 3.90 to 4.08 Å. The ligand’s torsion, similar to its six rotatable bonds, is proven in Fig. 11c. These outcomes underscore Quercetin’s sturdy binding functionality to EGFR throughout numerous power states with out inducing destabilizing structural alterations.
Earlier research using principal element evaluation (PCA) on protein-ligand molecular dynamics trajectories have offered invaluable insights into the structural determinants that govern ligand-induced results on molecular targets. This evaluation recognized important alterations in binding web site affinity, marked by pronounced vector variations inside the binding web site. These fluctuations are vividly illustrated within the protein’s secondary construction, with pink-highlighted vectors indicating the utmost fluctuations within the binding web site (Fig. 11d). Moreover, an brisk evaluation of mode 1 versus mode 2 revealed three distinct power minima, every fastidiously assessed for stability (Fig. 11e). Within the ESR1-Piperine (638024) complicated, the foundation imply sq. deviation (RMSD) ranged round 1.0 Å, whereas the radius of gyration (rGyr) prolonged between 4.75 and 5.25 Å. The torsion of the ligand, similar to its 4 rotatable bonds, is displayed in Fig. 11f. Regardless of these structural fluctuations, non-bonding interactions remained constant all through the simulation, highlighting the sustained binding efficacy of the ligand molecules.
The collective movement of the CYP19A1-Quercetin (5280343) complicated is visualized in Fig. 11g-i, the place the protein vectors on the interplay web site are oriented in a particular route, indicating a secure interplay (Fig. 11g). The general protein motion additionally follows a unidirectional sample, reinforcing the soundness of the complicated. The free power panorama (FEL) plot of the CYP19A1-Quercetin (5280343) complicated reveals three native power minima (Fig. 11h), reflecting a restricted variety of energetically favoured conformations, much like these noticed within the EGFR-Quercetin complicated. The Quercetin inside the CYP19A1-Quercetin (5280343) complicated reveals an RMSD vary of 0.8 Å, a radius of gyration (rGyr) between 3.90 and 4.08 Å, and torsion knowledge for its six rotatable bonds (Fig. 11i). These findings collectively assist the soundness of the ligand inside the complicated.
Publish molecular dynamics structural evaluation of protein-ligand complicated depicting (a) EGFR-Quercetin Protein Vectors, (b) EGFR-Quercetin PCA1 vs. PCA2 evaluation, (c) EGFR-Quercetin heatmap representing RMSD, (d) ESR1-Piperine Protein Vectors, (e) ESR1-Piperine PCA1 vs. PCA2 evaluation, (f) ESR1-Piperine heatmap representing RMSD, (g) CYP19A1-Quercetin Protein Vectors, (h) CYP19A1-Quercetin PCA1 vs. PCA2 evaluation, (i) CYP19A1-Quercetin heatmap representing RMSD.